Abstract
Atomic force microscopy (AFM) was used to study the morphological changes of two Gram-negative pathogens, Pseudomonas aeruginosa and Escherichia coli, after exposure to nitric oxide (NO). The time-dependent effects of NO released from a xerogel coating and the concentration-dependent effects rendered by a small-molecule that releases NO in a bolus were examined and compared. Bacteria exhibited irregular and degraded exteriors. With NO-releasing surfaces, an increase in surface debris and disorganized adhesion patterns were observed compared to controls. Analysis of cell surface topography revealed that increasing membrane roughness correlated with higher doses of NO. At a lower total dose, NO delivered via a bolus resulted in greater membrane roughness than NO released from a surface via a sustained flux. At sub-inhibitory levels, treatment with amoxicillin, an antibiotic known to compromise the integrity of the cell wall, led to morphologies resembling those resulting from NO treatment. Our observations indicate that cell envelope deterioration is a visible consequence of NO-exposure for both Gram-negative species studied.
Keywords: antimicrobial, atomic force microscopy, Gram-negative, morphology, nitric oxide
1. Introduction
Nitric oxide (NO) is a highly reactive diatomic radical endogenously produced by the enzyme-catalyzed oxidation of L-arginine to L-citrulline. It has been implicated as a mediator in multiple physiological processes, ranging from regulatory roles in the cardiovascular and nervous system to the inducible host response to infection.(1,2) Various therapeutic properties attributed to NO, including tumor cytotoxicity,(3,4) antimicrobial activity, and improved wound healing and tissue integration at implant sites,(5) may prove beneficial in a number of pharmacological applications.(6,7) Due to its reactivity, diverse regulatory roles, and short half-life in blood (< 1 sec),(8) the ability to target therapeutic NO delivery locally is critical. Nitric oxide donating compounds, such as N-diazeniumdiolates(6) and S-nitrosothiols,(9,10) decompose to release NO and hence serve as vehicles for its storage and transport. A number of materials, including nanoparticles,(11,12) films and coatings,(13,14) and small molecules,(15,16) have employed NO-donor chemistry with varied physicochemical and NO-release properties.
The role of NO in the innate immune response is a conserved feature through a wide range of species, from Drosophila to human.(17) In mammals, macrophages and other immune cells produce NO in response to invading pathogens.(18) The antimicrobial properties of NO may be elicited by direct modification of biomacromolecules or by formation of reactive nitrogen species (RNS) via reaction with oxygen (O2) or superoxide (O2−).(19) These RNS may render nitrosative stress by the formation of compounds such as dinitrogen trioxide (N2O3) and oxidative stress via the formation of peroxynitrite (ONOO−).(19-22) The spectrum of potential bactericidal mechanisms is thus broad, encompassing DNA damage resulting from deamination of deoxyribonucleotides, protein damage via numerous potential reactive sites (e.g. heme groups, thiols, amines) that disrupts normal cellular transport and metabolism, and membrane damage propagated by radical lipid peroxidation. The local physiological environment plays a key role in determining the metabolic pathways available to NO, and it would thus be expected that the bactericidal mechanism(s) of NO produced endogenously in phagosomal compartments would differ from NO released extracellularly (e.g., from an implanted biomaterial) as a result of differences in local conditions and substrates available in the biological milieu.
In vitro, NO has proven a potent antimicrobial agent effective against a range of microorganisms, including both Gram-negative and Gram-positive bacteria. Gaseous NO was found to be toxic against a number of pathogenic species, including Candida albicans and methacillin-resistant Staphylococcus aureus.(23) N-Diazeniumdiolate-modified NO-releasing surfaces have been shown to reduce initial Pseudomonas aeruginosa adhesion relative to controls,(24-26) and kill those that do adhere.(27) Nitric oxide release from silica nanoparticles has been characterized by significant toxicity to bacterial cells with reduced toxicity to L929 mouse fibroblasts.(28) While the bactericidal effects of NO and NO-releasing biomaterials have been demonstrated repeatedly, details on the primary targets resulting in bacterial cytotoxicity and the corresponding cellular effects of NO on microbial species remain speculative.
Morphological analyses of bacteria aid in understanding mechanisms of antibiotic action by allowing visualization of changes in the appearance of the microbe undergone subsequent to treatment. While electron microscopy has been employed toward this end for decades,(29-31) atomic force microscopy (AFM) has been used with increasing frequency.(32-37) As a surface characterization tool, AFM is ideal for morphological studies of surface-adhered bacteria as it allows cells to be imaged in situ with high resolution without requiring chemical drying, metal coating, or exposure to ultra-high vacuum. An added benefit of AFM is the flexible and adaptable nature of cantilevers as transducers that allow detection of other physical (e.g., elasticity) or chemical (e.g., charge distribution) surface parameters simultaneously with the acquisition of height information. Atomic force microscopy has been applied to visualizing the antimicrobial action of peptides,(32-34) chitosan,(35) quantum dots,(36) and the β-lactam antibiotics penicillin and amoxicillin.(37)
Herein, we report a morphological analysis of P. aeruginosa and Escherichia coli after exposure to NO released from two N-diazeniumdiolate-modified materials: a small molecule NO-donor derived from proline (PROLI/NO) and a NO donor-modified xerogel surface coating. The diazeniumdiolate moiety stores two molecules of the antimicrobial agent NO on each functionalized amine. Exposure to proton sources such as buffer and blood catalyzes the release of NO. Using topographical surface mapping and nanometer-scale height resolution, changes in bacteria shape and surface roughness were studied as a function of exposure time, material, and quantity of NO released.
2. Materials and methods
2.1 Materials
Ethanol and methanol were purchased from Fisher Scientific (Pittsburgh, PA). Argon, NO, nitrogen (N2), and a 25.7 ppm gaseous NO standard in N2 were purchased from National Welders (Raleigh, NC). N-(6-aminohexyl) aminopropyltrimethoxysilane (AHAP3) was obtained from Gelest (Tullytown, PA). Amoxicillin was obtained from Fluka (Buchs, Switzerland). Isobutyltrimethoxysilane (BTMOS), L-proline, sodium methoxide, and dimethylsulfoxide (DMSO) were purchased from Sigma-Aldrich (St. Louis, MO). The amino- and alkoxysilanes were stored over desiccant. The above chemicals were used without further purification. Distilled water was purified with a Millipore Milli-Q UV Gradient A-10 system (Bedford, MA) to a resistivity of 18.2 MΩ cm.
2.2 Cell culture P. aeruginosa
(ATCC #19143) and E. coli (ATCC #53323) were obtained from American Type Culture Collection (Manassas, VA) and cultured in tryptic soy broth (TSB). Stock cultures were prepared and stored at −80 °C for subsequent experiments. A 1-mL aliquot from an overnight culture was inoculated in ~100 mL of TSB and incubated at 37 °C for 3-5 h until the culture reached mid-exponential log phase as determined from optical density at 600 nm (OD600 = 0.2 ± 0.1), corresponding to ~108 colony forming units (cfu) mL−1.
2.3 Synthesis of xerogel films
Glass slides were coated with a 40% (v:v total silane content) AHAP3/BTMOS xerogel film via a 2-step process as described by Marxer et al.(13) Briefly, 120 μl BTMOS was mixed with 60 μl water, 200 μl ethanol, and 10 μl of 0.5 M HCl for 1 h. Then, 80 μl of AHAP3 was added, and the solution was mixed for an additional hour. Glass slides were cut into sections (dim. 13 × 17.5 mm), rinsed with ultrapure water and ethanol, dried under a stream of nitrogen, and cleaned for 30 min in a UV-ozone cleaner (BioForce, Ames, IA). To cast a film, 40 μl of the sol was pipetted onto clean glass slides, dried for 30 min at ambient temperature, and cured at 85 °C for 3 d. Control xerogel films were stored in desiccators at 22 °C.
2.4 NO-donor synthesis and characterization
Xerogels were modified to release NO by exposing the films to 5 atm of NO for 72 h as previously described.(13) The NO chamber was flushed twice with 5 atm Ar to remove atmospheric impurities (e.g., oxygen, water) prior to introducing NO gas. After 3 d, unreacted NO was removed by flushing the vessel with Ar.
L-proline was converted to PROLI/NO following a procedure previously reported by Saavedra, et al.(15) Briefly, 10 g of L-proline was dissolved in 39 mL of 25% sodium methoxide in methanol. This solution was combined with an additional 20 mL of methanol, and exposed to NO gas (5 atm) as described above. The resulting PROLI/NO formed as a white precipitate that was collected via filtration, washed with ether, and vacuum dried. All NO-releasing materials (i.e., PROLI/NO and xerogels) were stored in vials purged with nitrogen at −20 °C until use in order to stabilize the NO donor.
A chemiluminescent nitric oxide analyzer (NOA) (Sievers Model 280, Boulder, CO) was used to measure NO release in real time. A known quantity of the NO-release material was placed in a flask containing phosphate buffered saline (PBS, pH = 7.4) positioned in a water bath maintained at 37 °C. The NO generated via diazeniumdiolate decomposition was carried into the analyzer via a stream of N2 bubbled into the solution at a flow rate of 80 mL min−1. The detector was calibrated by a 2-point curve using an atmospheric sample passed through an NO zero filter and a 25.7 ppm NO standard.
2.5 Bacterial adhesion to control and NO-releasing xerogels
Prior to use, the NO-releasing xerogels were allowed to reach ambient temperature. Control and NO-releasing films were rinsed briefly with ultrapure water and dried under a stream of nitrogen immediately prior to bacterial adhesion. After diluting bacterial suspensions in TSB (1:2 in PBS), a 200-μL aliquot of the solution was beaded directly onto the xerogel surface. Substrates were covered to reduce evaporation and incubated for either 1 h (in preparation for treatment in antibiotic solutions) or 2 h (NO-releasing xerogels) at 37 °C to allow for bacterial adhesion.
2.6 Antimicrobial treatment
For time points exceeding 2 h, NO-releasing xerogels with adhered bacteria were placed in vials containing 5 mL of PBS and incubated at 37 °C for the remainder of the exposure period. For treatment with PROLI/NO, the appropriate mass was first weighed into chilled, dry vials. The correct volume of PBS (~5 mL) was added to obtain the desired concentration, vortexed briefly, and a control (unmodified) xerogel with adhered, untreated bacteria was immediately added to the PROLI/NO solution and incubated for 2 h at 37 °C. Amoxicillin treatment was achieved by preparing a 1 μg mL−1 solution in PBS from a 9.6 mg mL−1 stock solution in DMSO, into which a control xerogel (with adhered bacteria) was placed and incubated for 2 h at 37 °C. Each xerogel time point and antibiotic concentration was replicated for each species studied. A detailed description of the substrate rinsing process is provided below.
2.7 Substrate preparation technique
A gentle-rinse procedure was developed to ensure that adhered cells remained relatively undisturbed for subsequent treatment and/or imaging experiments. A rinse step was accomplished by pipetting a single, 300-μL aliquot of the appropriate rinse solution onto the substrate. Immediately thereafter, the solution was removed with a narrow-tipped pipette in 100-μL increments. Of note, the pipette was only applied to the outer edge of the substrate to remove residual rinse solution so as not to disturb the surface to be imaged.
For both control and NO-releasing xerogels, excess cell suspension in TSB was removed from the surface after the adhesion period, followed by five consecutive rinses with PBS to remove TSB, non-adhered cells, and trace cellular components. If adhered cells were subsequently treated, the substrate was added to a vial containing the appropriate concentration of antimicrobial agent in PBS (PROLI/NO, amoxicillin) for 2 h or PBS (40% AHAP3/BTMOS xerogel time points exceeding 2 h) at 37 °C. The final rinse steps consisted of 5 washes with ultrapure water to remove antibiotic and/or salts, after which the substrate was dried using capillary action by placing absorbent paper at the edge of the substrate to draw excess water off the surface. A thin layer of water remained, which was allowed to evaporate prior to imaging. hawse have previously shown that the process of drying bacteria at xerogel surfaces does not affect their viability.(38)
2.8 AFM Imaging
Simultaneous AFM height, amplitude, and phase images were obtained in AC mode on the air-dried substrates using an Asylum MFP-3D AFM (Santa Barbara, CA). Olympus AC240TS silicon beam cantilevers (Center Valley, PA) with a spring constant of 2 N m−1 and resonant frequency of 70 kHz were used to image bacteria in air. At least three 20 μm2 survey images were obtained at random locations at an interior region of each substrate. Additional images captured control and antibiotic treated cells at greater magnifications. Images were acquired at a resolution of 512 × 512 pixels and scan speed of 1 Hz. Individual root-mean-square (rms) roughness of cell membranes was calculated using the MFP-3D software from 500 and 800 nm2 regions of P. aeruginosa and E. coli, respectively. Images used for roughness determination were acquired in the central part of a cell and were flattened by one order to reduce contributions from cell curvature at the edges of the image. Membrane roughness values were averaged from at least 3 different cells per species, agent, and dose. Artificial color and light were added to the three-dimensional reconstructions of height data to aid visualization of image detail.
3. Results and Discussion
3.1 Material characterization and experimental design
Bacterial cells were exposed to two NO-releasing materials that differ in their kinetic release profiles. A representative NO-release profile and the average integrated dose delivered from a 40% AHAP3/BTMOS xerogel film as a function of time are provided in Figure 1 and Table 1, respectively. Nitric oxide is released from these films via a surface flux, increasing rapidly upon exposure to aqueous solution and reaching a maximum within half an hour. Thereafter, the NO release gradually reduces to a flux averaging ~50 pmol cm−2 s−1 over 24 h. Hetrick, et al. reported that a total dose of NO between 375 and 425 nmol cm−2 delivered from 40 % AHAP3/BTMOS xerogels 5 – 7 h after initial bacterial adhesion was sufficient to eradicate all adhered P. aeruginosa cells at room temperature.(27) By contrast, PROLI/NO is a water-soluble, small molecule diazeniumdiolate NO donor derived from the amino acid proline. PROLI/NO releases a bolus of nitric oxide upon breakdown by water. Due to a short half-life (t1/2 = 100 s), the majority of stored NO (10.4 ± 2.1 μmol NO mg−1) is released within 300 s (Table 1). Standard plating experiments indicated a minimum bactericidal concentration of 4 and 8 mg mL−1 PROLI/NO after 2 hours (MBC120) for E. coli and P. aeruginosa, respectively (3-log reduction in cfu).
Figure 1.
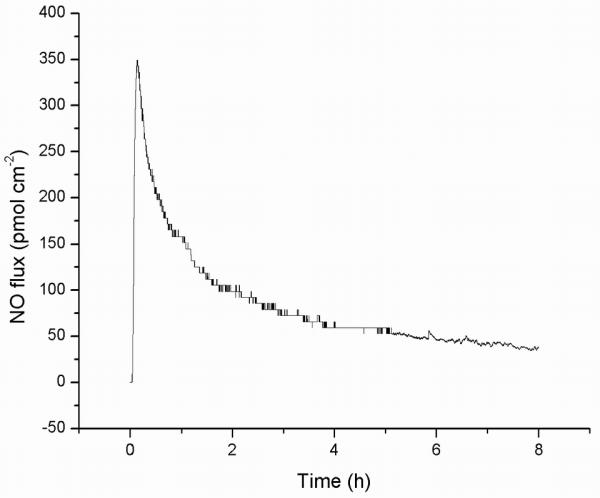
Nitric oxide released from a 40% AHAP3/BTMOS xerogel coating in PBS at 37 °C.
Table 1.
Measured NO released from antibacterial materials
| material | NO delivery | exposure | dose NOb |
|---|---|---|---|
| xerogel | surface flux | 2 h | 1.32 ±0.13 (μmol cm−2) |
| 4 h | 1.93 ±0.19 | ||
| 6 h | 2.39 ± 0.23 | ||
| 8 h | 2.70 ± 0.27 | ||
|
| |||
| PROLI/NO | bolus | 70 s | 3.4 ± 1.1 (μmol mg−1) |
| 300 sa | 8.8 ± 2.0 | ||
The majority of NO has been released after this time. The total NO released by 1 mg of PROLI/NO is 10.4 ± 2.1 μmol NO.
Average dose of NO reported with standard deviation
Nitric oxide release from the materials used in this study was measured in deoxygenated PBS per the convention for measuring NO accurately via chemiluminescence. In contrast, the experiments to evaluate the bactericidal effects of these materials were conducted in normal PBS (i.e., not deoxygenated), since the bacteria are aerobic. Nevertheless, control experiments indicated that the NO release from AHAP3/BTMOS xerogels in normal PBS was indistinguishable from measurements made in deoxygenated PBS over 0 – 24 h, the time frame of our experiments.
Due to the nature of diazeniumdiolate decomposition to NO (at physiological pH and temperature), bacterial adhesion to NO-releasing xerogel films occurs concurrently with exposure of the bacteria to a local NO flux. Although slower compared to control xerogels, the surface coverage of P. aeruginosa to NO-releasing 40% AHAP3/BTMOS xerogels have been shown to reach a steady state at 60 min under static conditions.(27) For these experiments, NO-releasing surfaces were exposed to cell suspensions for 2 h, followed by removal of loose cells and preparation of substrates for imaging (for 2 h time points) or transfer of the substrates to PBS (for extended time points). To apply a similar approach for treatment with antibiotic solutions (i.e., PROLI/NO and amoxicillin), bacteria were allowed to adhere to control xerogel substrates for an hour prior to transfer of the cell-covered substrate to a vial of antibiotic solution. Treating surface-adhered cells with antibacterial agents provided the added benefits of capturing cell damage and debris locally on the surface at the time of treatment while more nearly approximating an infection-causing scenario.
3.2 Morphologies of P. aeruginosa and E. coli adhered to control xerogels
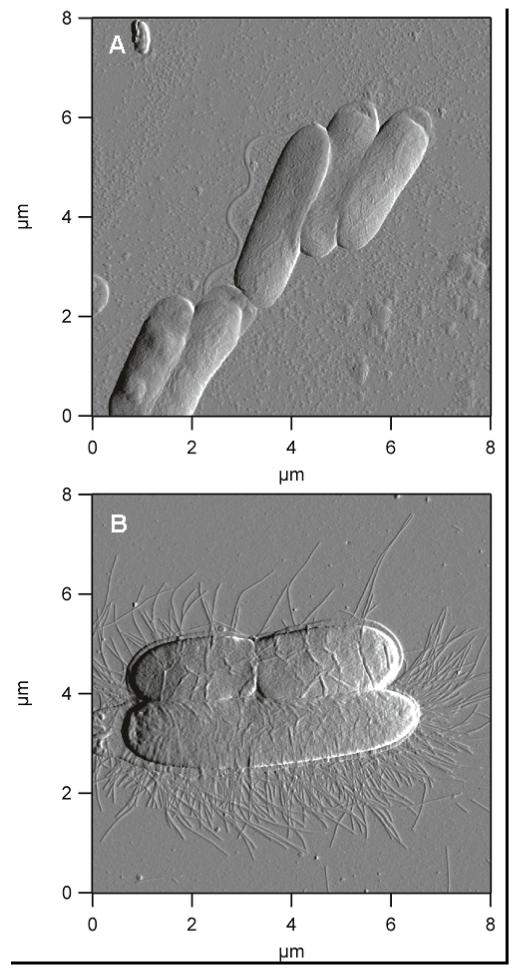
Control 40% AHAP3/BTMOS xerogels have been reported to be non-toxic to bacteria adhered at exposure periods > 24 h.(38) Thus, they represent a suitable substrate for the study of normal morphologies, while providing a consistent sub-stratum for comparison of healthy and antibiotic-treated (solution) cells to those treated via a surface flux of NO. Representative images of untreated P. aeruginosa and E. coli cells adhered to control xerogels are illustrated in Figure 2. P. aeruginosa cells are rod-shaped and exhibit regular dimensions (~ 1 × 3 μm) with an inflated appearance and smooth cell exterior. Also rod-shaped, E. coli are somewhat larger and more variable in length. On control surfaces, P. aeruginosa adhered in well organized patterns that maximized contact along the long axis of the cells while maintaining apparent structural integrity. By comparison, E. coli cells tended to maintain some physical separation (i.e., adhering individually or in small groups of 2 to 3 cells). While P. aeruginosa are motile via flagella, E. coli have a characteristic crown of fimbriae. Healthy cells of both species appeared intact with no visible pores, holes, grooves, or breakages in the cell envelope. To negate any possible effect derived from the presence of proline, E. coli and P. aeruginosa preadsorbed on control xerogels were incubated for 2 h with 2 and 4 mg mL−1 proline, respectively. Physically, the proline exposed cells were indistinguishable from untreated cells. Equivalent cell viability was verified by growing colonies on nutrient agar.
Figure 2.
Deflection images depict the morphology of healthy (A) P. aeruginosa; and (B) E. coli bacteria.
3.3 Morphologies of NO-treated P. aeruginosa and E. coli
Sub-bactericidal concentrations such as those used in this study have historically been applied to study antimicrobial effects as they provide ‘snapshots’ of the organism’s morphology between healthy and dead states.(31,33,39) Figure 3 illustrates examples of P. aeruginosa and E. coli bacteria after exposure to NO. A representative collection of images was chosen to demonstrate the full spectrum of related morphologies resulting from NO treatment. Many of the effects of NO exposure (e.g., membrane degradation) were exhibited at multiple doses. A comprehensive list of the morphologies observed after treatment with NO is included in Table 2. Many bullets are identical or similar for each species indicating related mechanisms of action against both Gram-negative bacteria. Such features include mild to extensive membrane degradation, debris present on the surface in the vicinity of cells, blebbing or cellular parts abnormally attached to cells, cellular collapse, and lysis. Spheroplast formation and increasingly short cellular length were more infrequent morphologies common to both species, generally observed on xerogels after longer exposures to surfaces fluxes of NO (6 – 8 h, Fig. 3D-E). Neither species exhibited population arrangements on NO-releasing xerogels similar to those observed on controls. P. aeruginosa adhered in a disorganized array, while E. coli cells abandoned their active tendency to maintain spatial separation on a surface. Cells were occasionally observed to adhere across a previously adhered cell, despite a low overall surface coverage.
Figure 3.
Three-dimensional reconstructions of height images illustrate P. aeruginosa and E. coli morphologies, shown in the left and right columns, respectively. Parts (A-E) depict cells treated by NO flux from 40% AHAP3/BTMOS xerogels for (A, C) 4 h; (B) 6 h; (D, E) 8 h. Parts (F-J) show examples of bacteria treated with sub-bactericidal concentrations of PROLI/NO for 2 h at (F-H) 1 mg mL−1; (I) 4 mg mL−1; and (J) 2 mg mL−1
Table 2.
Morphologies exhibited by Gram-negative bacteria after NO treatment
| P. aeruginosa | E. coli |
|---|---|
|
|
The collection of morphologies observed indicates cell envelope damage as a visible and significant contributing mechanism to the cytotoxic effect of NO against P. aeruginosa and E. coli. The morphologies observed in this AFM study closely resemble those reported by Li, et al. who concluded that their antibacterial peptides disrupted, permeabilized, and eventually destroyed the stability of the outer and inner lipid membranes of P. aeruginosa and E. coli.(33) The single morphology that points strictly to protein damage in this study, the broken fimbriae frequently observed for E. coli, also occurred at the cell membrane. Nitric oxide-mediated membrane damage has recently been reported by Hetrick, et al. using confocal fluorescence microscopy.(28) In that study, P. aeruginosa membranes became permeable to propidium iodide, a fluorescent dye that may only enter bacterial cells with compromised membranes, after exposure to NO-releasing silica nanoparticles (maximum NO flux ≈ 21700 ppb mg−1).
Both delivery routes used in this study released NO exterior to, but in the vicinity of, surface-adhered bacteria. As a broad-spectrum antibiotic, NO exerts both oxidative and nitrosative stress on biomolecules at cell surfaces. Physically, NO and O2 are lipophilic and membrane permeable leading to the concentration and sequestering of these molecules and their reactive metabolites (e.g., N2O3) near lipid bilayers,(21) where membrane-bound and other local proteins become targets of nitrosative stress. The formation of peroxynitrite from the reaction of NO with intracellular O2− initiates the radical peroxidation of lipid membranes (oxidative stress), potentially the cause of the observed degradation of these structural components.
3.4 Analysis of membrane roughness after NO treatment
To further characterize the effect of NO on the Gram-negative cell, AFM images were obtained of the bacterial membrane. The cell membrane damage by NO was readily observed in representative three-dimensional reconstructions from height images after 2, 4, 6, and 8 h exposures to NO flux from xerogels (Figure 4). The range of the scales was held constant (15 nm) to allow comparison between AFM images. Gram-negative cell envelopes are composed of an outer and inner lipid membrane, each about 10 nm thick, separated by a thin, cross-linked peptidoglycan layer. Holes in the outer lipid membrane began forming as early as 2 h (Fig. 4B) after exposure to NO surface fluxes. Over time, the degree of membrane degradation continued to increase, with large holes and crevices penetrating into the inner membrane (Fig 4C-E). In fact, at exposure times exceeding 4 h the roughness of the membrane exceeded the height scale. The image analysis software was then used to apply light/shadow at identical angles and pitch to add a sense of depth. By comparison, Figure 4A depicts the membrane of a healthy P. aeruginosa cell adhered to a control xerogel for 24 h. Its morphological analysis confirms that changes in membrane roughness are a function of NO-exposure and not surface residence time.
Figure 4.
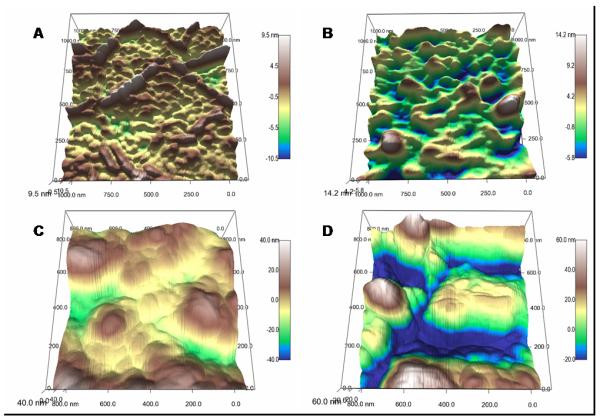
P. aeruginosa membranes on a control surface (A), and after exposure to NO surface flux for (B) 2 h, (C) 4 h, (D) 6 h, and (E) 8 h from a 40% AHAP3/BTMOS xerogel, and (F) after exposure to 0.07 μM NO in solution (bolus delivery).
The quantitative rms roughness of cell membranes as a function of bacteria species and NO exposure time/concentration is given in Table 3. As expected, longer exposure to a NO surface flux (40% AHAP3/BTMOS xerogels) correlates with rougher cell membranes ranging from ~1.6 nm for controls to nearly 4 and 12 nm after 8 h NO release for P. aeruginosa and E. coli, respectively. Similarly, membrane roughness was greater for E. coli for cells treated with greater concentrations of PROLI/NO (15.2 ± 4.6 and 28.9 ± 7.9 nm for 1 and 2 mg mL−1, respectively) (Fig. 5). Of note, the measured roughness for P. aeruginosa was the same at both subbactericidal (MBC120 of PROLI/NO for P. aeruginosa is 8 mg mL−1) concentrations of PROLI/NO (2 and 4 mg mL−1), and approximately double that measured for the longest exposure to NO-releasing xerogels. If the deterioration of the lipid bilayer leads to increased surface roughness, this may be indirect evidence that protein and/or DNA damage contributes significantly to the cytotoxic effect of PROLI/NO against this species.
Table 3.
Root-mean-square (rms) roughness of cell membranes
| Species | material | exposure | average (nm)a |
|---|---|---|---|
| P. aeruginosa | xerogel | control | 1.61 ±0.27 |
| 2 h | 2.13 ±0.09 | ||
| 4 h | 2.25 ±0.30 | ||
| 6 h | 3.20 ± 0.31 | ||
| 8 h | 3.84 ± 0.76 | ||
|
|
|||
| PROLI/NO | 2 mg mL−1 | 7.15 ±1.52 | |
| 4 mg mL−1 | 7.10 ±0.86 | ||
| NO solution | 0.07 μM | 3.16 ±0.61 | |
|
| |||
| E. coli | xerogel | control | 1.55 ±0.21 |
| 2 h | 2.66 ± 0.66 | ||
| 4 h | 3.24 ± 0.45 | ||
| 6 h | 4.33 ± 0.55 | ||
| 8h | 11.7 ± 1.9 | ||
|
|
|||
| PROLI/NO | 1 mg mL−1 | 15.2 ±4.6 | |
| 2 mg mL−1 | 28.9 ± 7.9 | ||
Average rms roughness reported with standard deviation
Figure 5.
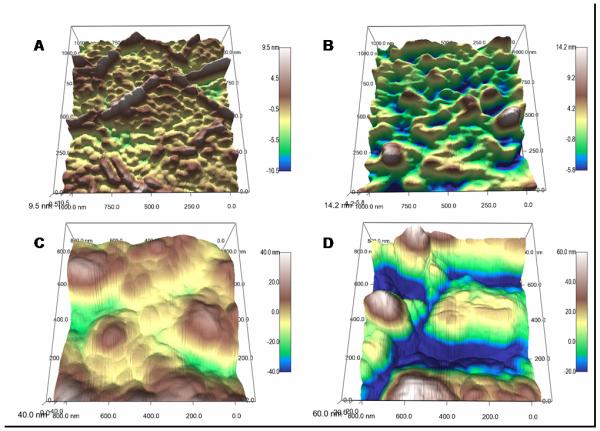
Three-dimensional reconstructions of E. coli membranes compare untreated (A) relative to the degradation sustained after 6 h NO-release from a xerogel surface (B). Treatment with sub-bactericidal PROLI/NO concentrations of 1 mg mL−1 (C) and 2 mg mL−1 (D) show more extensive damage only 2 h after exposure. Note the difference in scale range.
3.5 Comparison of morphologies resulting from NO and amoxicillin treatment
Amoxicillin, a β-lactam antibiotic, functions by inhibiting enzymes that cross-link chains in the peptidoglycan layer. Treatment of E. coli with a sub-bactericidal concentration of amoxicillin was therefore expected to generate morphologies typical of cell wall degradation. (P. aeruginosa was not treated with amoxicillin as it has demonstrated resistance to the effects of this β-lactam.) The most common morphologies observed by imaging amoxicillin-treated E. coli were perforations in the cell surface (pore formation) and regions of collapsed cell wall, both concentrated (but not restricted to) the apical ends of the cells (Fig. 6C, D). These observations are in agreement with a study that used AFM to compare the morphological changes sustained by E. coli after treatment with amoxicillin and its parent molecule, the natural product penicillin.(37) Both E. coli (Fig. 6A) and P. aeruginosa (Fig. 6B) exhibited analogous morphologies to the amoxicillin-treated cells when treated with low levels of NO. In fact, these morphologies were only visualized on NO-releasing xerogels at 2 h time points, and thus at the lowest concentrations of NO treatment used in this study. At greater NO doses, these morphologies were obscured as the extent of membrane damage increased and the incidence of localized effects decreased. Interestingly, the morphologies observed for treatment of E. coli with penicillin, described by Yang, et al.(37) as randomly distributed grooves and holes, strongly resemble the morphological changes sustained by both E. coli and P. aeruginosa at greater NO doses. The similarity of morphologies observed after treatment with amoxicillin, which functions by a known mechanism of action, to those after exposure to NO offers additional support to the conclusion that exposure to NO results in the deterioration of the cell envelope of Gram-negative bacteria.
Figure 6.
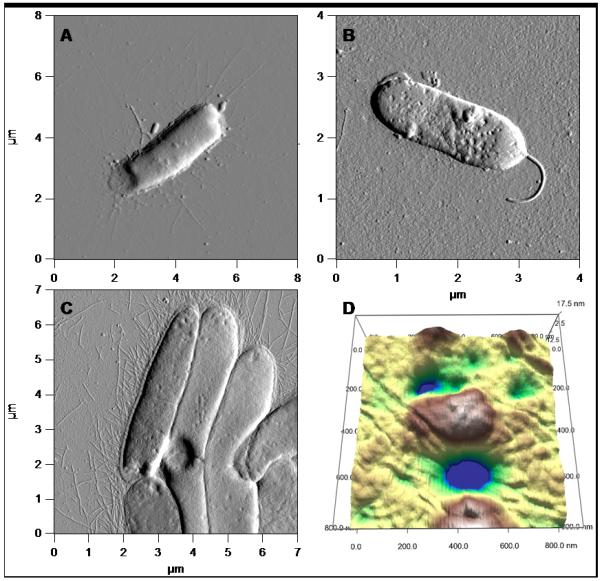
Comparison of the morphological effects caused by treatment with NO on E. coli (A) and P. aeruginosa (B) strongly resemble the morphologies demonstrated by E. coli after treatment with amoxicillin (C), a β-lactamase that inhibits cell wall synthesis. Furthermore, the three-dimensional rendering of the compromised cell wall (D) after treatment with amoxicillin resembles the holes and crevices exhibited by P. aeruginosa and E. coli after NO treatment, indicative of membrane damage.
3.6 Comparison of NO delivery methods
Comparing NO-delivery methods proved less straightforward than varying exposure time or concentration for a single delivery method. Both the NO-release kinetics (slow versus fast) and manner of delivery (surface flux versus burst of NO release) differ for 40% AHAP3/BTMOS xerogels and PROLI/NO. While treatment with PROLI/NO resulted in rougher, more highly deteriorated cells than exposure to a sustained surface flux as visualized qualitatively (Fig. 3) and measured quantitatively (Table 3), the total amount of NO released as a bolus from sub-bactericidal concentrations of PROLI/NO exceeded that delivered by sub-bactericidal fluxes of NO from 40% AHAP3/BTMOS. As the molecules of NO released from PROLI/NO were dispersed throughout the solution rather than being concentrated at the location of bacterial adhesion (i.e., at the surface), this may explain the significantly greater quantities of NO necessary induce a bactericidal effect.
To deconvolute the efficacy of the delivery routes, P. aeruginosa cells were treated with a bolus of NO slightly less than that delivered by a 40% AHAP3/BTMOS xerogel over 2 h. Xerogel-adhered P. aeruginosa cells were immediately added to a 0.07 μM solution of NO in 5 mL of PBS (prepared from a saturated NO solution) and incubated for 2 h under conditions identical to the PROLI/NO experiments. The membrane roughness remained greater when cells were treated by a bolus compared to surface flux from an NO-releasing 40% AHAP3/BTMOS xerogel (3.2 ± 0.6 nm vs. 2.1 ± 0.1 nm, respectively) (Table 3), and a comparison of the P. aeruginosa membrane after treatment with an NO solution (Fig. 4F) to that after 2 h exposure to surface flux (Fig. 4B) clearly demonstrates larger pores and crevices in the former.
Bacterial species have evolved strategies to protect against the harmful effects of NO.(40) For example, specific transcription factors (e.g., SoxR, OxyR) identified in E. coli are capable of sensing NO released from macrophages and respond by up-regulating gene expression to combat toxic effects.(41,42) Once detected in vivo, these proteins convert NO to less toxic by-products such as nitrate.(43) Although the molecules of NO released from PROLI/NO are dispersed throughout the medium, the total concentrations of NO and RNS available to react with the adhered cells are greater during the initial period of incubation when they are released via a bolus. Delivered in a highly concentrated burst, NO may devastate bacterial cells before they are capable of mounting a defense. As an extension of this hypothesis, future studies should investigate the behaviour of bacteria selected after exposure to increasing concentrations of NO in an attempt to foster tolerance and probe the upper limit of resistance to NO.
Conclusions
The morphologies of two Gram-negative species of bacteria were observed using AFM after treatment with the antimicrobial agent NO. Quantitative measurements of surface roughness and qualitative observation of increased surface debris and changes in cell shape (e.g., blebbing) and adhesion patterns indicate that membrane degradation is a significant contributing factor to NO’s bacterial cytotoxicity. Comparison of morphological effects perpetrated via a single, known mechanism (i.e., inhibition of cell wall synthesis by amoxicillin) to those observed from treatment of NO aids in confirming the antimicrobial mechanism of the latter. By evaluating NO sources with different NO-release kinetics, we conclude that greater levels of NO released over short durations are more damaging to Gram-negative bacteria than sustained, lower-level surface fluxes. The double lipid bilayer of Gram-negative bateria typically acts as a permeability barrier to antibiotics that function within the cell. Ironically, it is this same structural characteristic that renders these cells particularly susceptible to NO-induced membrane damage. As degradation of the cell envelope leads to an increase in permeability, treatment with NO may elicit synergistic effects when used in concert with antibiotics.
Acknowledgements
The authors gratefully acknowledge research support from the National Institute of Health (NIH EB000708).
References
- 1.Lowenstein CJ, Dinerman JL, Snyder SH. Nitric oxide: A physiologic messenger. Ann Intern Med. 1994;120(3):227–237. doi: 10.7326/0003-4819-120-3-199402010-00009. [DOI] [PubMed] [Google Scholar]
- 2.Bogdan C. Nitric oxide and the immune response. Nat Immunol. 2001;2:907–916. doi: 10.1038/ni1001-907. [DOI] [PubMed] [Google Scholar]
- 3.Bonavida B, Khineche S, Huerta-Yepez S, Garbán H. Therapeutic potential of nitric oxide in cancer. Drug Resist Updat. 2006;9(3):157–173. doi: 10.1016/j.drup.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 4.Buga GM, Ignarro JL. Nitric Oxide and Cancer. Academic Press; San Diego: 2000. pp. 895–920. [Google Scholar]
- 5.Hetrick EM, Prichard HL, Klitzman B, Schoenfisch MH. Reduced foreign body response at nitric oxide-releasing subcutaneous implants. Biomaterials. 2007;28:4571–4580. doi: 10.1016/j.biomaterials.2007.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keefer LK. Progress toward clinical application of the nitric oxide-releasing diazeniumdiolates. Annu Rev Pharmacol Toxicol. 2003;43:585–607. doi: 10.1146/annurev.pharmtox.43.100901.135831. [DOI] [PubMed] [Google Scholar]
- 7.Napoli C, Ignarro LJ. Nitric oxide-releasing drugs. Annu Rev Pharmacol Toxicol. 2003;43:97–123. doi: 10.1146/annurev.pharmtox.43.100901.140226. [DOI] [PubMed] [Google Scholar]
- 8.Smith DJ, Chakravarthy D, Pulfer S, Simmons ML, Saavedra JE, Davies KM, Hutsell TC, Mooradian DL, Hanson SR, Keefer LK. Nitric oxide-releasing polymers containing the [N(O)NO]-group. J Med Chem. 1996;39:1148–1156. doi: 10.1021/jm950652b. [DOI] [PubMed] [Google Scholar]
- 9.Lyn D, Williams H. The chemistry of S-nitrosothiols. Acc Chem Res. 1999;32:869–876. [Google Scholar]
- 10.Butler AR, Rhodes P. Chemistry, analysis, and biological rolse of S-Nitrosothiols. Anal Biochem. 1997;249:1–9. doi: 10.1006/abio.1997.2129. [DOI] [PubMed] [Google Scholar]
- 11.Shin JH, Metzger SK, Schoenfisch MH. Synthesis of nitric oxide-releasing silica nanoparticles. J Am Chem Soc. 2007;129:4612–4619. doi: 10.1021/ja0674338. [DOI] [PubMed] [Google Scholar]
- 12.Shin JH, Schoenfisch MH. Inorganic/organic hybrid silica nanoparticles as a nitric oxide delivery scaffold. Chem Mater. 2008;20:239–249. doi: 10.1021/cm702526q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marxer SM, Rothrock AR, Nablo BJ, Robbins ME, Schoenfisch MH. Preparation of nitric oxide (NO)-releasing sol-gels for biomaterial applications. Chem Mater. 2003;15:4193–4199. [Google Scholar]
- 14.Robbins ME, Hopper ED, Schoenfisch MH. Synthesis and characterization of nitric oxide-releasing sol-gel microarrays. Langmuir. 2004;20:10296–10302. doi: 10.1021/la048368n. [DOI] [PubMed] [Google Scholar]
- 15.Saavedra JE, Southan GJ, Davies KM, Lundell A, Markou C, Hanson SR, Adrie C, Hurford WE, Zapol WM, Keefer LK. Localizing antithrombotic and vasodilatory activity with a novel, ultrafast nitric oxide donor. J Med Chem. 1996;39:4361–4365. doi: 10.1021/jm960616s. [DOI] [PubMed] [Google Scholar]
- 16.Hrabie JA, Klose JR, Wink DA, Keefer LK. New nitric oxide-releasing zwitterions derived from polyamines. J Org Chem. 1993;58:1472–1476. [Google Scholar]
- 17.Foley E, O’Farrell PH. Nitric oxide contributes to induction of innate immune responses to gram-negative bacteria in Drosophila. Genes Dev. 2003;17:115–125. doi: 10.1101/gad.1018503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.MacMicking J, Xie QW, Nathan C. Nitric oxide and macrophage function. Annu Rev Immunol. 1997;15:323–350. doi: 10.1146/annurev.immunol.15.1.323. [DOI] [PubMed] [Google Scholar]
- 19.Wink DA, Mitchell JB. Chemical biology of nitric oxide: Insights into regulatory, cytotoxic, and cytoprotective mechanisms of nitric oxide. Free Radic Biol and Med. 1998;25(4):434–456. doi: 10.1016/s0891-5849(98)00092-6. [DOI] [PubMed] [Google Scholar]
- 20.Fang FC. Mechanisms of nitric oxide-related antimicrobial activity. J Clin Invest. 1997;99:2818–2825. doi: 10.1172/JCI119473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moller MN, Li Q, Lancaster JRJ, Denicola A. Acceleration of nitric oxide autooxidation and nitrosation by membranes. Life. 2007;59:243–248. doi: 10.1080/15216540701311147. [DOI] [PubMed] [Google Scholar]
- 22.Zaki MH, Akuta T, Akaike T. Nitric oxide-induced nitrative stress involved in microbial pathogenesis. J Pharmacol Sci. 2005;98:117–129. doi: 10.1254/jphs.crj05004x. [DOI] [PubMed] [Google Scholar]
- 23.Ghaffari A, Miller CC, McMullin B, Ghahary A. Potential application of gaseous nitric oxide as a topical antimicrobial agent. Nitric Oxide. 2006;14:21–29. doi: 10.1016/j.niox.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 24.Nablo BJ, Chen T-Y, Schoenfisch MH. Sol-gel derived nitric-oxide releasing materials that reduce bacterial adhesion. J Am Chem Soc. 2001;123:9712–9713. doi: 10.1021/ja0165077. [DOI] [PubMed] [Google Scholar]
- 25.Nablo BJ, Rothrock AR, Schoenfisch MH. Nitric oxide-releasing sol-gels as antibacterial coatings for orthopedic implants. Biomaterials. 2005;26:917–924. doi: 10.1016/j.biomaterials.2004.03.031. [DOI] [PubMed] [Google Scholar]
- 26.Nablo BJ, Schoenfisch MH. Antibacterial properties of nitric oxide-releasing sol-gels. J Biomed Mater Res. 2003;67A:1276–1283. doi: 10.1002/jbm.a.20030. [DOI] [PubMed] [Google Scholar]
- 27.Hetrick EM, Schoenfisch MH. Antibacterial nitric oxide-releasing xerogels: Cell viability and parallel plate flow cell adhesion studies. Biomaterials. 2007;28:1948–1956. doi: 10.1016/j.biomaterials.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 28.Hetrick EM, Shin JH, Stasko NA, Johnson BA, Wespe DA, Holmuhamedov E, Schoenfisch MH. Bactericidal efficacy of nitric oxide-releasing silica nanoparticles. ACS Nano. 2007;2:235–246. doi: 10.1021/nn700191f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Waisbren SJ, Hurley DJ, Waisbren BA. Morphological expression of antibiotic synergism against Pseudomonas aeruginosa as observed by scanning electron microscopy. Antimicrob Agents Chemother. 1980;18(6):969–975. doi: 10.1128/aac.18.6.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klainer AS, Russell RRB. Effect of the inhibition of protein synthesis on the Escherichia coli cell envelope. Antimicrob Agents Chemother. 1974;6(2):216–224. doi: 10.1128/aac.6.2.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iida K, Koike M. Cell wall alterations of Gram-negative bacteria by aminoglycoside antibiotics. Antimicrob Agents Chemother. 1974;5(1):95–97. doi: 10.1128/aac.5.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Da Silva A, Teschke O. Effects of the antimicrobial peptide PGLa on live Escherichia coli. Biochim Biophys Acta. 2003;1643:95–103. doi: 10.1016/j.bbamcr.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 33.Li A, Lee PY, Ho B, Ding JL, Lim CT. Atomic force microscopy study of the antimicrobial action of Sushi peptides on Gram-negative bacteria. Biochim Biophys Acta. 2007;1768:411–418. doi: 10.1016/j.bbamem.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 34.Meincken M, Holroyd DL, Rautenbach M. Atomic force microscopy study of the effect of antimicrobial peptides on the cell envelope of Escherichia coli. Antimicrob Agents Chemother. 2005;49(10):4085–4092. doi: 10.1128/AAC.49.10.4085-4092.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eaton P, Fernandes JC, Pereira E, Pintado ME, Malcata FX. Atomic force microscopy study of the antibacterial effects of chitosans on Escherichia coli and Staphylococcus aureus. Ultramicroscopy. 2008;108(10):1128–1134. doi: 10.1016/j.ultramic.2008.04.015. [DOI] [PubMed] [Google Scholar]
- 36.Lu Z, Li CM, Bao H, Qiao Y, Toh Y, Yang X. Mechanism of antimicrobial activity of CdTe quantum dots. Langmuir. 2008;24:5445–5452. doi: 10.1021/la704075r. [DOI] [PubMed] [Google Scholar]
- 37.Yang L, Wang K, Tan W, He X, Jin R, Li J, Li H. Atomic force microscopy study of different effects of natural and semisynthetic β-lactam on the cell envelope of Escherichia coli. Anal Chem. 2006;78:7341–7345. doi: 10.1021/ac0604890. [DOI] [PubMed] [Google Scholar]
- 38.Deupree SM, Schoenfisch MH. Quantitative method for determining the lateral strength of bacterial adhesion and application for characterizing adhesion kinetics. Langmuir. 2008;24:4700–4707. doi: 10.1021/la800417g. [DOI] [PubMed] [Google Scholar]
- 39.Hayami H, Goto T, Kawahara M, Ohi Y. Activities of β-lactams, fluoroquinolones, amikacin, and fosfomycin alone and in combination against Pseudomonas aeruginosa isolated from complicated urinary tract infections. J Infect Chemother. 1999;5:130–138. doi: 10.1007/s101560050022. [DOI] [PubMed] [Google Scholar]
- 40.Crawford MJ, Goldberg DE. Role for the Salmonella flavohemoglobin in protection from nitric oxide. J Biol Chem. 1998;273(20):12543–12547. doi: 10.1074/jbc.273.20.12543. [DOI] [PubMed] [Google Scholar]
- 41.Nunoshiba T, DeRojas-Walker T, Wishnok JS, Tannenbaum SR, Demple B. Activation by nitric oxide of an oxidative-stress response that defends Escherichia coli against activated macrophages. Proc Natl Acad Sci. 1993;90:9993–9997. doi: 10.1073/pnas.90.21.9993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hausladen A, Privalle CT, Keng T, DeAngelo J, Stamler JS. Nitrosative stress: Activation of the transcription factor OxyR. Cell. 1996;86:719–729. doi: 10.1016/s0092-8674(00)80147-6. [DOI] [PubMed] [Google Scholar]
- 43.Hausladen A, Gow AJ, Stamler JS. Nitrosative stress: Metabolic pathway involving the flavohemoglobin. Proc Natl Acad Sci. 1998;95:14100–14105. doi: 10.1073/pnas.95.24.14100. [DOI] [PMC free article] [PubMed] [Google Scholar]