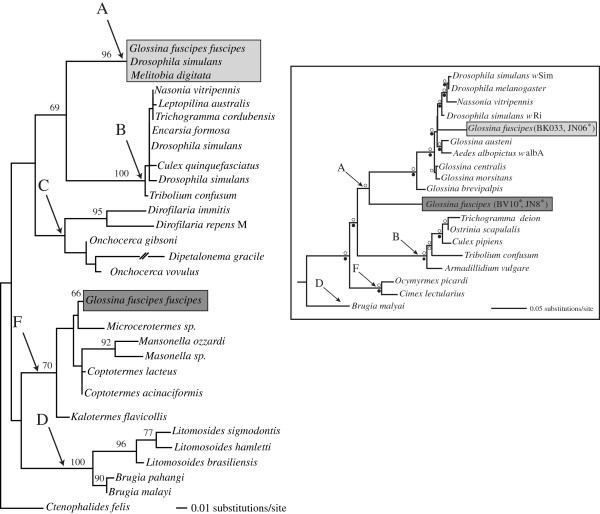
Figure 2.
Rooted maximum likelihood gene tree for groEL and the tree inferred from four MLST loci (inset). Arrows point to the branch that leads to each supergroup (A-F) and are labeled accordingly for both phylogenies. Shaded clades indicate the position of G. f. fuscipes Wolbachia sequences. Light gray shading shows the position of Group 1 sequences within supergroup A. Dark gray shading shows the position of Group 2 sequences. Individual hosts that belong to each clade are shown in the additional file. (See Additional file 1: Table S1 and Figure S1). The groEL tree includes Wolbachia strains from Glossina fuscipes fuscipes and representatives from most known supergroups. For the groEL tree, the outgroup was selected arbitrarily to reflect relationships in [25]. Numbers above branches are bootstrap support percentages that are above 65%. Within supergroups for groEL, bootstrap support was below 65% and thus not reported. Inset (Upper right): A generalized version of the MLST phylogeny (See Additional file 1: Figure S1). An open circle above the branch indicates a Bayesian posterior probability of 1.0 and a closed circle below the branch is a bootstrap of 90% and above. The shaded boxes correspond to the shading of branches in the groEL phylogeny. Individuals analyzed for both groEL and MLST are indicated with an *.