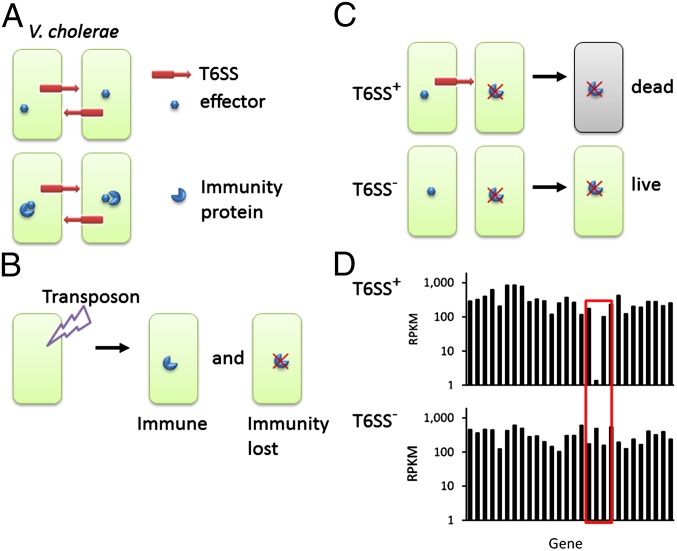
Fig. 1.
Schematic diagram of the selection method for immunity genes. (A) T6SS organelles of two V. cholerae sister cells actively attack each other and inject a potentially lethal effector protein. Because a cognate immunity protein binds to and neutralizes the effector, the cells survive the attack. A corollary to this model is that injection of the effector protein per se should not be a lethal event in sister cell–sister cell interactions. (B) Saturating transposon mutagenesis generates two populations of mutants, one still immune and the other lost immunity due to transposon inactivation of a T6SS effector–immunity protein. (C) Mutants that lost immunity were killed by T6SS of neighboring cells and diminished in the mutant pool. However, such mutants would survive if constructed in a strain lacking T6SS apparatus. (D) Deep sequencing of transposon insertion junctions (Tn-seq) is used to quantify the relative abundance of transposon insertions in every gene in the context of the T6SS+ and T6SS− backgrounds. The genes for T6SS immunity proteins correspond to those genes that show a lower normalized value of mapped DNA sequence reads expressed as RPKM in the T6SS+ strain than in the T6SS− strain.