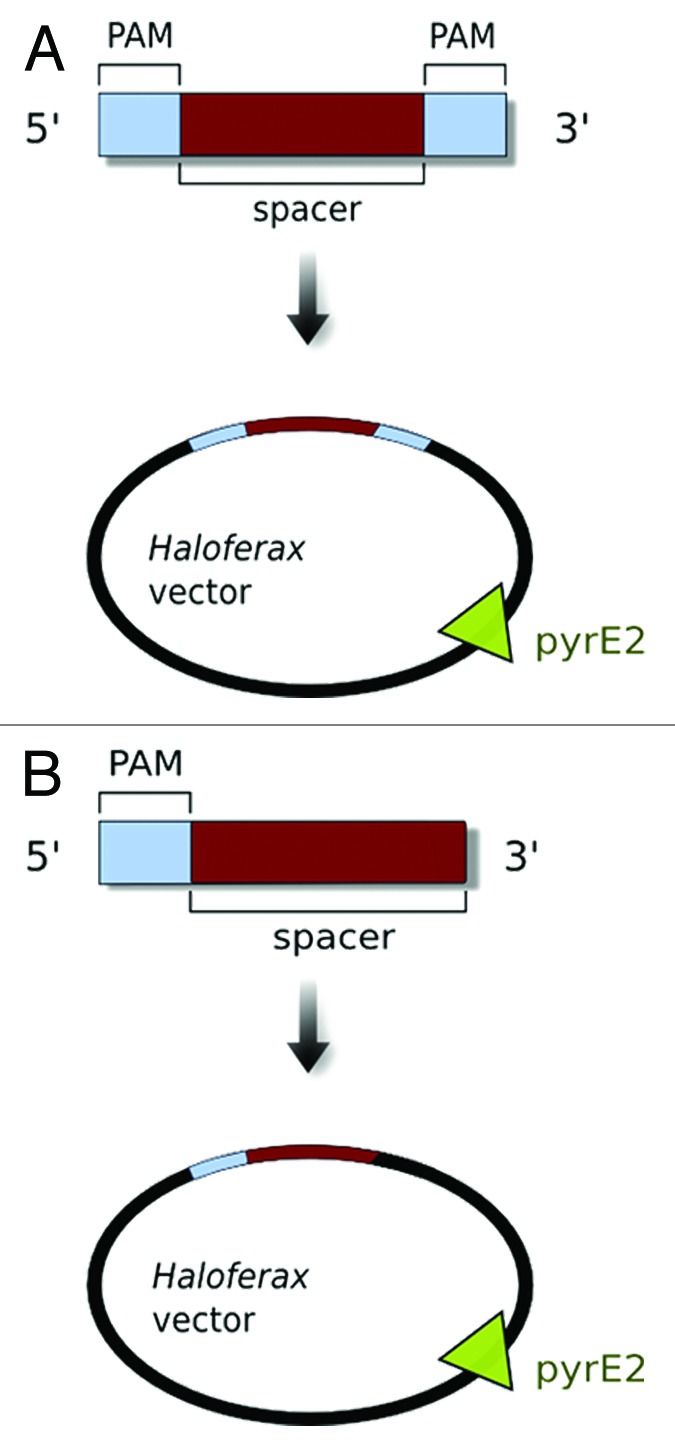
Figure 3. An artificial invader for Haloferax. To challenge the Haloferax defense system, we generated an artificial invader consisting of a spacer sequence (from one of the Haloferax CRISPR loci, shown in red) and an adjacent sequence with all possible two- and three-nucleotide combinations as potential PAM sequences (shown in light blue). These were cloned into a Haloferax plasmid vector that also carried a selection marker pyrE2 (which makes growth of the ΔpyrE2-Haloferax recipient strain independent of supplied uracil). (A) Initial experiments were performed with potential PAM sequences up- and downstream of the spacer sequence. (B) PAM localization experiments showed that the PAM sequence is only required upstream of the spacer sequence and thus we positioned PAM sequences upstream only.
