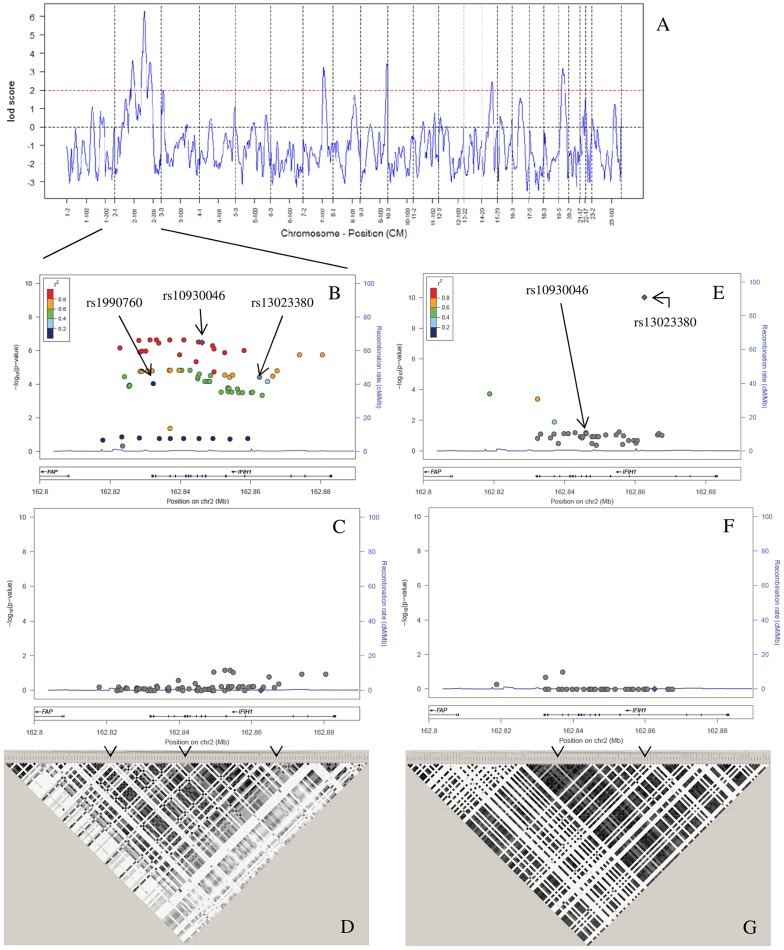
Figure 2. Admixture mapping and conditional analysis.
(A) A whole-genome admixture scan on AA SLE cases identified 7 admixture signals that achieved the predefined LOD score >2 (red dashed line). (B) We performed an imputation-based association analysis of IFIH1, which was identified as the most promising candidate gene by a case-control study on 20 candidate genes in the largest peak (2q22–24), followed by 4-SNP haplotype conditional analysis (C). Filled dots indicate the −log10 P values for the association to SLE, and color coding represents inter-marker correlation (r2) between the strongest associated SNP, rs10930046 (“purple diamond”), and the individual SNPs, as shown in the color bar. (C) After conditioning the 4-marker haplotypes for the three markers rs1990760–rs10930046–rs13023380, all individual SNP associations are explained as shown. (D) We analyzed the LD between SNPs on the ImmunoChip, and these LD values were used as a reference panel for imputation in AA. Darker color denotes higher correlation between markers (r2). The LD pattern showed high correlation between markers, making it possible to increase SNP density by imputation. The three independently associated SNPs identified in (B) are denoted by arrows. (E) We performed an imputation based case-control association analysis in EA. Filled dots indicate the −log10 P values for each SNP, and color coding represents the inter-marker correlation (r2) between each individual SNP and the strongest associated SNP, rs13023380 (“purple diamond”), as shown in the color bar. (F) We then performed a two SNP haplotype analysis followed by a three marker haplotype analysis conditioned on the two independent variants rs10930046 and rs13023380. (G) LD analysis of SNPs on the ImmunoChip reference panel showed low inter-marker correlation, which largely precluded imputation based association. Darker color indicates greater r2. Arrows indicate the position of the independent SNPs.