Fig. 3. An LNA gapmer causes degradation of targeted DEADSouth mRNA.
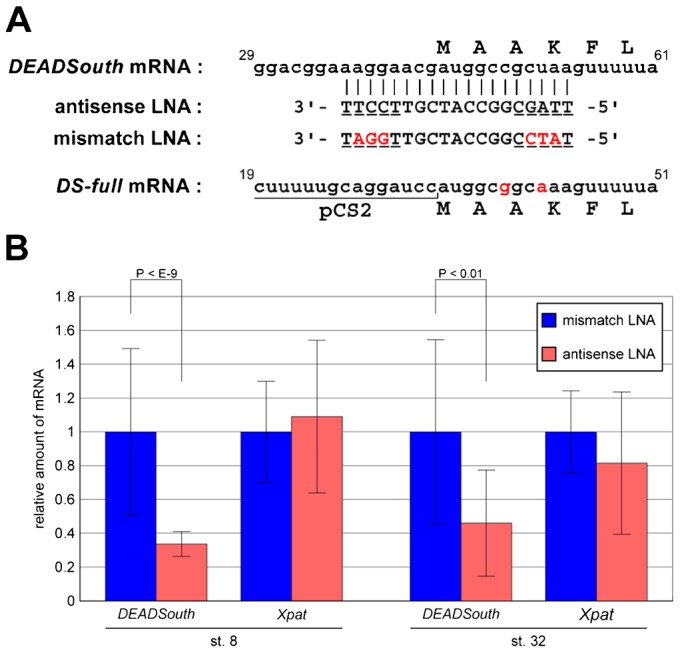
(A) Sequence alignment of DEADSouth mRNA (nucleotide position 29–61, accession no. AF190623), antisense and mismatch LNA gapmers and synthesized DEADSouth mRNA (DS-full). Upper letters indicate the amino acid sequence. LNA modifications and mismatch bases are underlined and in red, respectively. The sequence of DS-full mRNA was predicted from that of the template DNA. (B) Quantification of DEADSouth and Xpat mRNAs in stage 8 and 32 embryos injected with LNA gapmers. The amounts of these mRNAs were determined by real-time RT-PCR, normalized to EF1α mRNA levels as an internal control, and shown as relative amounts to the mRNA level in embryos injected with mismatch LNA gapmer. P-values and s.d. are indicated.
