Figure 1.
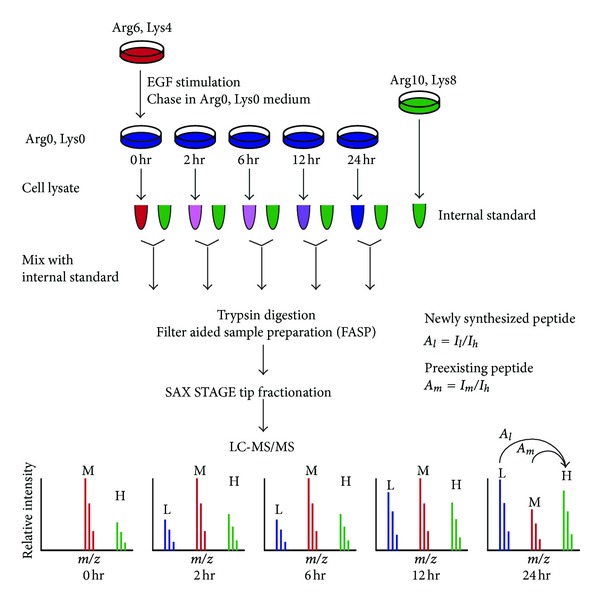
Using internal standard-assisted synthesis and degradation mass spectrometry (iSDMS) to study the roles of EGF on global protein synthesis and degradation. SKBR3_shVPS4B and the parental SKBR3 cells were first labeled with 13C6-arginine (Arg6) and D4-lysine (Lys4) medium (labeled in red). After overnight serum starvation, Arg6/Lys4-labeled cells were stimulated with 100 ng/mL EGF in medium supplemented with regular arginine (Arg0) and lysine (Lys0) for 0, 2, 6, 12, and 24 hr (labeled in blue). Protein isolated from SKBR3_shVPS4B cells labeled with 13C6 15N4-arginine (Arg10) and 13C6 15N2-lysine (Lys8)—internal standard (labeled in green)—was spiked into each sample at a ratio of 1 : 3 (wt/wt). The mixtures were digested by the Filter Aided Sample Preparation (FASP) procedure, followed by strong anion exchange (SAX) peptide fractionation. Peptides were analyzed by online LC-MS/MS using an LTQ-Orbitrap mass spectrometer. The relative abundance of the newly synthesized (A l) or preexisting peptides (A m) was defined as the ratio of mass spectrometric peak intensities of the unlabeled peptides (I l) or Arg6/Lys4-labeled peptides (I m) to the intensities of the Arg10/Lys8-labeled peptides (I h), respectively.
