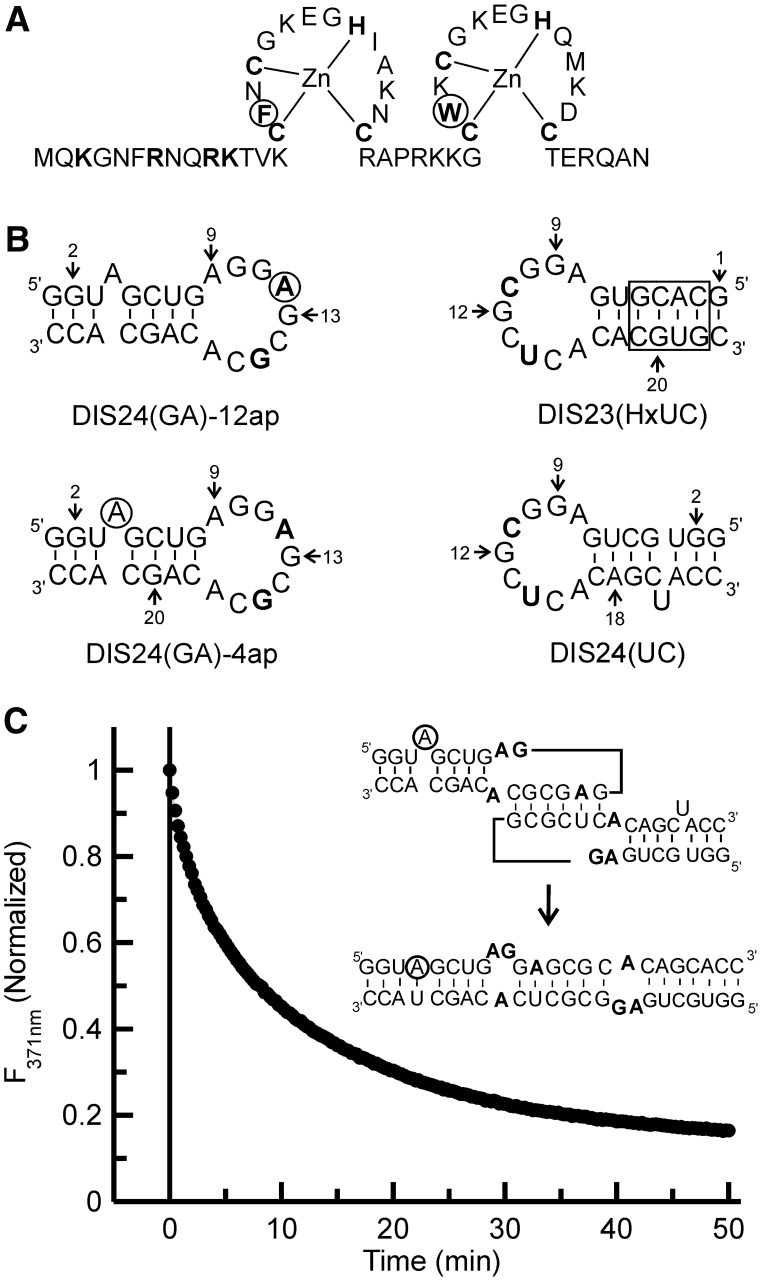
Figure 1.
(A) NCp7 wild-type protein sequence. Zn coordinating and basic residues in the N-terminus are shown in bold. The aromatic residues Phe16 and Trp37 are in bold and circled. (B) SL1 hairpin constructs. The residues changed from the wild-type sequence in the DIS loop region are bolded, and the adenines that are replaced by 2-AP in the two fluorescent constructs [DIS24(GA)-4ap and DIS24(GA)-12ap] are circled. The DIS24(UC) contains a complementary bulged uracil that forms a base-pair on maturation and the exchanged stem in the DIS23(HxUC), which disfavors formation of the duplex via strand exchange, are boxed. The DIS23(GA) hairpin used in the NMR experiments is the same sequence as the DIS24(GA) constructs with the exception that the bulged adenosine at position 4 is deleted. (C) Representative plot of the DIS kissing complex to extended duplex conversion measured by 2-AP fluorescence emission quenching as a function of time after the addition of wild-type NCp7 to the DIS24(GA)-4ap•DIS24(UC) kissing complex at physiological pH (225 nM NCp7 protein was added to 100 nM of RNA kissing complex). (Inset) Schematic of the DIS kissing complex to extended duplex conversion is shown with the 2-AP position circled.