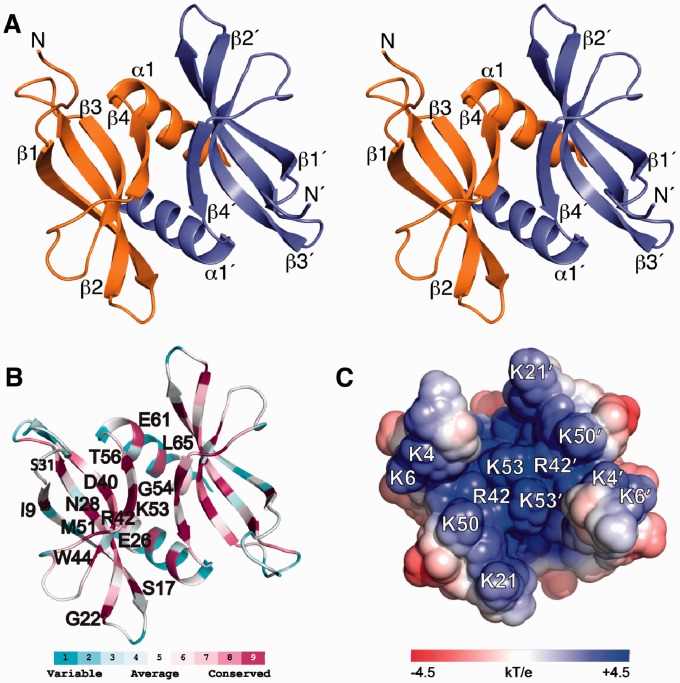
Figure 1.
Solution NMR structure of L. lactis apo-YdbC shown in the identical top-view orientation. (A) Stereoview of dimeric YdbC with labelled secondary structure elements and amino termini. (B) ConSurf (29,30) amino acid conservation mapped onto the lowest energy NMR structure. Highly conserved residues are labelled on the protein backbone of a single protomer. (C) Solvent exposed electrostatic potential (32) mapped onto the surface of apo-YdbC. Only the ssDNA-binding epitope is shown for clarity.