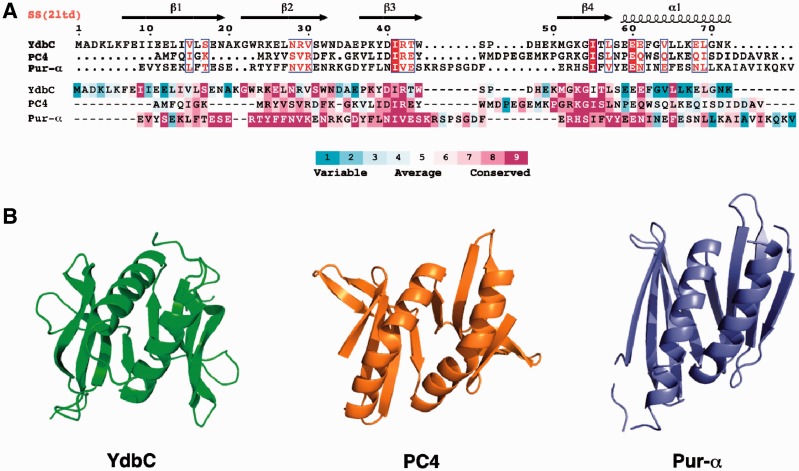
Figure 2.
(A) Structure-based sequence alignment (26,27) of L. lactis YdbC (DUF2128; PF09901), H. sapiens PC4 (PF02229) and B. burgdorferi PUR-α (DUF3276; PF11680). (Top) Sequence alignment rendered by ESPript (42) using default parameters for residue similarity calculations, where boxed residues represent identical (red box, white character) and similar (red character) amino acid conservation. (Bottom) Sequence alignment rendered using ConSurf (29,30) where residue conservation across individual protein domain families range from highly conserved (magenta) to variable (cyan). (B) Comparison of the solution NMR structure of L. lactis YdbC with crystal structures structurally similar apo-forms of dimeric ssDNA-binding proteins, H. sapiens PC4 (PDB ID: 1pcf) (43) and B. burgdorferi PUR-α (PDB ID: 3nm7) (8).