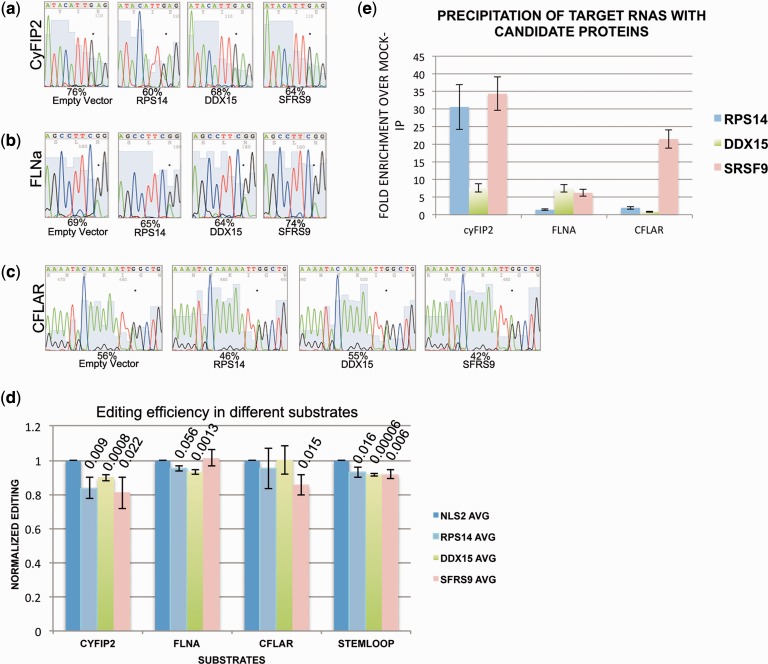
Figure 3.
Editing of endogenous substrates is selectively inhibited by repressors of editing. Editing of endogenous (a) CyFip2, (b) FLNa and (c) CFLAR RNA was determined on transfection of cDNAs encoding the three candidate repressors of editing RPS14, DDX15 and SFRS9. CyFIP2 editing is most strongly affected by expression RPS14 and SFRS9. FLNA, in contrast, is barely affected by any of the three candidates. CFLAR editing, finally, is only affected by SFRS9. Editing efficiency was calculated by dividing A and G peak height by G peak height. The edited adenosine is marked by an asterisk. (d) The average editing levels of three biological replicates are calculated and normalized to mock-transfected samples. P-values are calculated and indicated where significant. (e) myc tagged candidates were precipitated and the associated RNAs isolated. The presence of substrate RNAs in the precipitate was quantified by quantitative RT-PCR and normalized to the amount of RNA present in the cellular lysate. Fold enrichment was calculated by comparison to a mock IP with protein A sepharose beads only. cyFIP2 RNA is strongly enriched in IPs with SRSF9 and RPS14, while CFLAR RNA is strongly enriched with SFRS9 only.