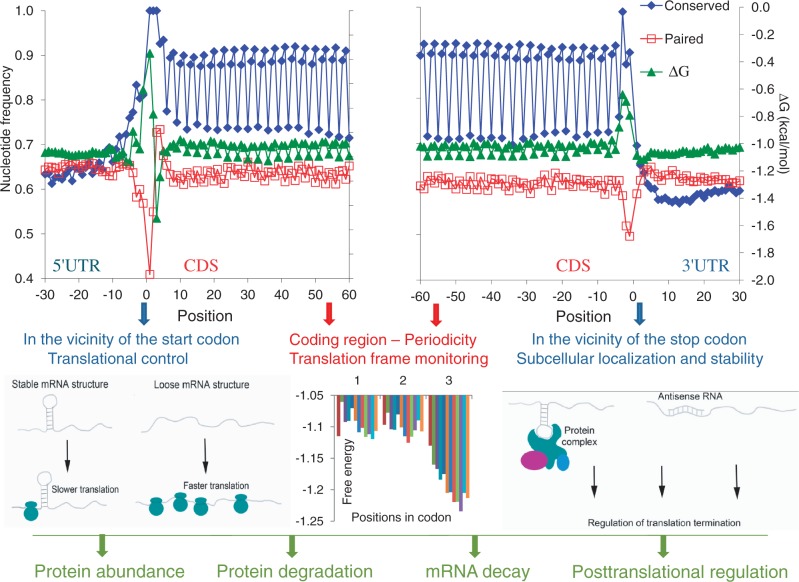
Figure 1.
Periodic pattern of nucleotide involvement in secondary structure formation and sequence conservation around the start and the stop codons in human mRNAs. Positions from −30 to −1 correspond to 5′-UTRs and positions from 1 to 60 correspond to CDSs (upper left panel). Positions from −60 to −1 correspond to CDSs and positions from 1 to 30 correspond to 3′-UTRs (upper right panel). Blue, sequence conservation in 6919 orthologous human and mouse mRNAs. Red, base-paired nucleotides in 19 317 human mRNAs. Green, free Gibbs energy of base-pairing in 19 317 human mRNAs. Structural features of the untranslated regions (UTRs) and coding sequences (CDSs) have a major role in the control of mRNA translation. The relaxed secondary structures in UTRs are common for many mRNAs and involved in regulation of initiation (low left panel) and termination (low right panel) of translation. Periodic pattern in the CDS is likely responsible for translation frame monitoring (low center panel). Groups of genes with distinct levels of expression (114) are presented in different colors. This figure is adapted from (10).