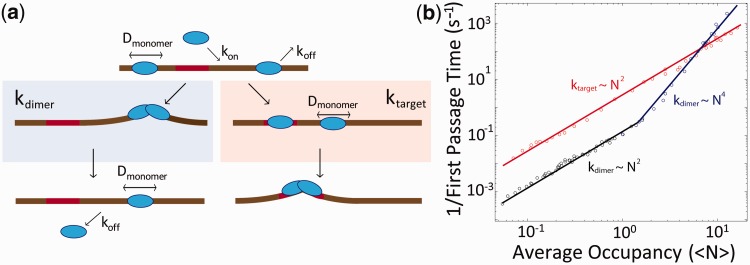
Figure 6.
Kinetics of TelK-induced dimerization and target search. (a) Model of TelK target search. TelK monomers in solution bind DNA with rate kon and scan rapidly along non-target DNA with mean diffusion coefficient Dmonomer = 1.8 µm2/s. Monomers localize the target site with rate ktarget and bind tightly or dissociate from non-target DNA with average rate koff = 0.24 s−1. Preferential and stable binding of a TelK monomer allows a second monomer to dimerize at the target site and form a kinked DNA–TelK complex primed for catalysis. Occasionally, mobile monomers encounter each other on non-target DNA with rate kdimer and form stable, immobile dimers (Ddimer < 1 × 10−4 µm2/s) that ‘test’ their substrate by condensing the DNA transiently. Eventually, dimers on non-target DNA dissociate or separate into mobile monomers again (rate < 0.01 s−1). (b) Implementation of kinetic model in (a) via stochastic simulations using experimentally derived kinetic parameters. Average rates for first dimerization kdimer (blue, black), and first target localization ktarget (red) as a function of average protein occupancy, <N>, on DNA. For mean occupancies >1, target-finding and dimerization rates obey simple scaling laws of N2 and N4, respectively. For mean occupancies <1, dimerization follows of N2 scaling. Provided TelK occupancy is reasonably small, target localization consistently occurs faster than dimerization. Simulated rates are in good agreement with experimentally derived rates of first dimerization.