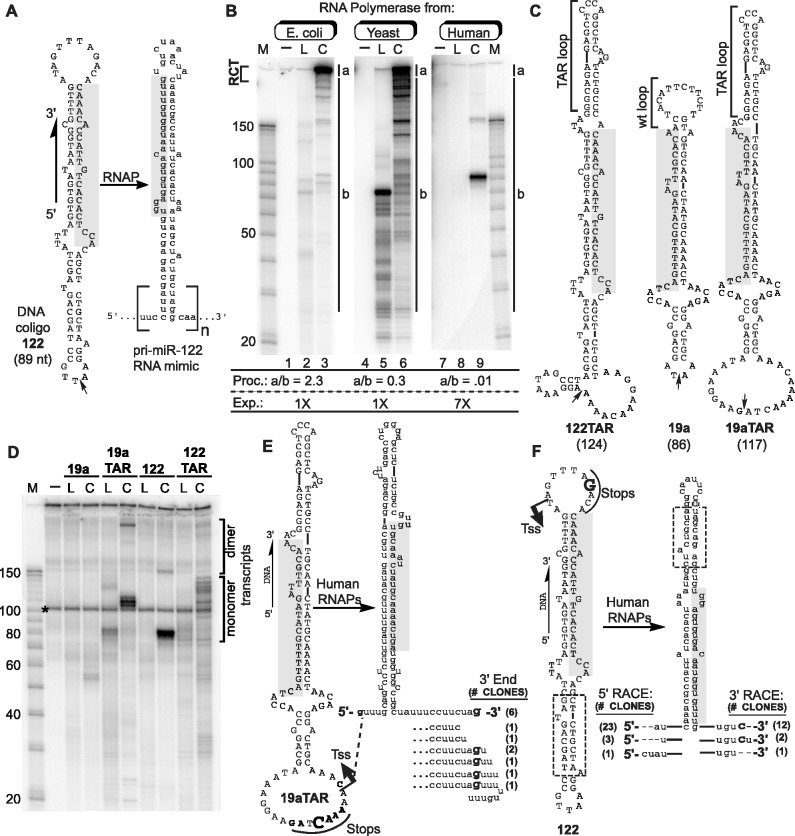
Figure 1.
Transcription of coligos by RNAPs. (A) Schematic illustration showing the sequence and predicted secondary structure of a coligo and its RNA. Coligo 122 was taken from the template strand of the human gene encoding miR-122. The subscript n denotes number of tandem repeats. RCT produces n » 1; n ∼ 1 indicates single round transcription leading to monomer transcripts. Small arrow: circularization site, linear forms are discontinuous here with 5′ phosphate. Shaded region, mature miR-122 or its cDNA. nt, size in nucleotides. (B) IVT of coligo 122 by RNAPs of varying evolutionary age and complexity. E. coli RNAP, yeast RNAP II and human RNAPs from HEK293T WCE were used for IVT. RNA was visualized on a denaturing polyacrylamide gel (DPAGE) by uniform [α-32P]-UTP incorporation. L, linear; C, coligo. Lanes 1, 4, 7: no template. M, RNA marker. Relative processivity (Proc.) defined by the a/b ratio, and relative exposure (Exp., i.e. Phosphorimager grayscale setting difference) are indicated. (C) Sequences and predicted secondary structures of coligos 122TAR, 19a and 19aTAR. (D) IVT using HEK293T WCE followed by DPAGE analysis of uniformly labeled transcripts. Dimer transcripts read through the termination site one time to produce tandem dimer transcripts. (E) Sequence analysis of transcripts made by human WCE from coligo template 19aTAR. The 19aTAR IVT products were isolated and their cDNA sequenced. Tss, transcription start site. (F) Sequence analysis of transcripts made by human WCE from coligo template 122. The 122 IVT product was isolated and sequenced using 5′ and 3′ RLM-RACE protocols.