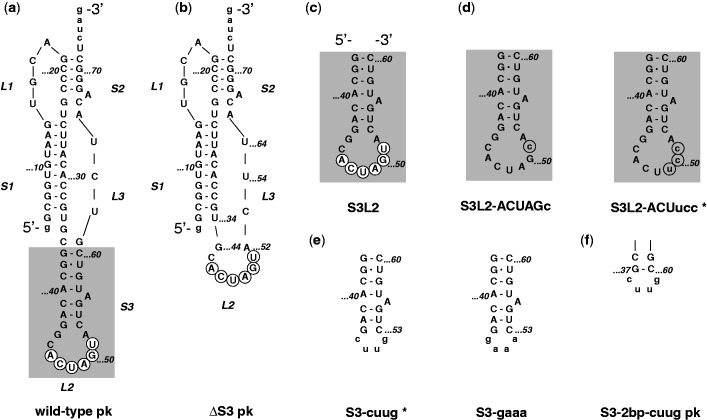
Figure 1.
SARS constructs. (a) The three-stemmed wild-type SARS pseudoknot. Stems are labeled S1, S2 and S3 in the order that they occur 5′ to 3′ along the RNA. Accordingly, loops are labeled L1, L2 and L3. Note that L1 and L3 join adjacent stems, while L2 closes S3 (highlighted using gray box). Only the last two digits of the wild-type sequence numbering are used for clarity. The palindromic sequence 5′-ACUAGU-3′ embedded into L2 is indicated using white circles. Dashes represent Watson–Crick and the dot G•U Wobble base-pairing as confirmed by NMR spectroscopy. (b) Stem 3 deletion mutant ΔS3 pk. (c) S3L2 hairpin construct S3L2 spanning nucleotides G37 to C60. (d) S3L2 hairpin constructs S3L2-ACUucc and S3L2-ACUAGc with L2 mutations that render the palindromic sequence asymmetrical (highlighted using gray circles) while conserving a Serine codon. (e) S3L2 hairpin constructs S3-cuug and S3-gaaa where the 9 nt L2 is replaced with the smaller tetraloops 5′-cuug-3′ and 5′-gaaa-3′, respectively. (f) SARS pseudoknot variants S3-2 bp-cuug pk with a shortened Stem 3 is capped with a 5′-cuug-3′ tetraloop. S3-cuug and S3L2-ACUucc variant constructs highlighted with an asterisk (*) were also generated in the context of full-length pk.