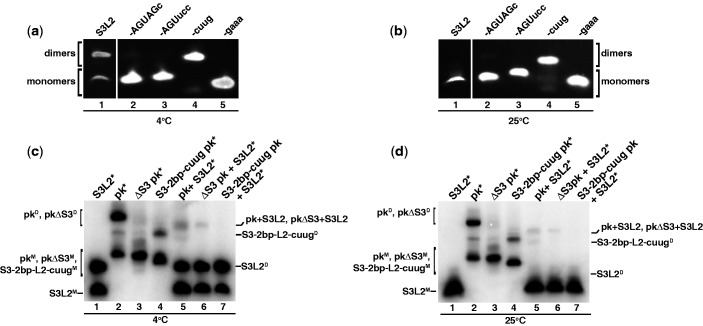
Figure 3.
Detection of SARS RNA dimers. RNA transcripts (150 µM/each) were incubated at 37°C for 30 min in the presence of 200 mM KCl without MgCl2. Samples were separated on a 10% native PAGE. (a) Native gel analysis at 4°C of S3L2-only variants: S3L2, S3L2-ACUAGc, S3L2-ACUucc, S3-cuug and S3-gaaa. Gels were stained with ethidium bromide and visualized by UV. S3L2 transcript was independently analysed. (b) Same as in (a) but at 25°C. (c) Native gel analysis at 4°C of the following transcripts: wild-type S3L2, wild-type pk, Stem 3 deletion mutant ΔS3 pk and S3-2 bp-cuug. RNA variants were incubated with 32P-labeled transcripts in trace [1 pM] indicated by *. (d) Same as in (c), but at 25°C. For (c) and (d) samples were separated on a 10% native PAGE, which was dried and analysed by phosphorimaging. Uppercase ‘M’ denotes monomer and uppercase ‘D’, dimer.