Figure 3.
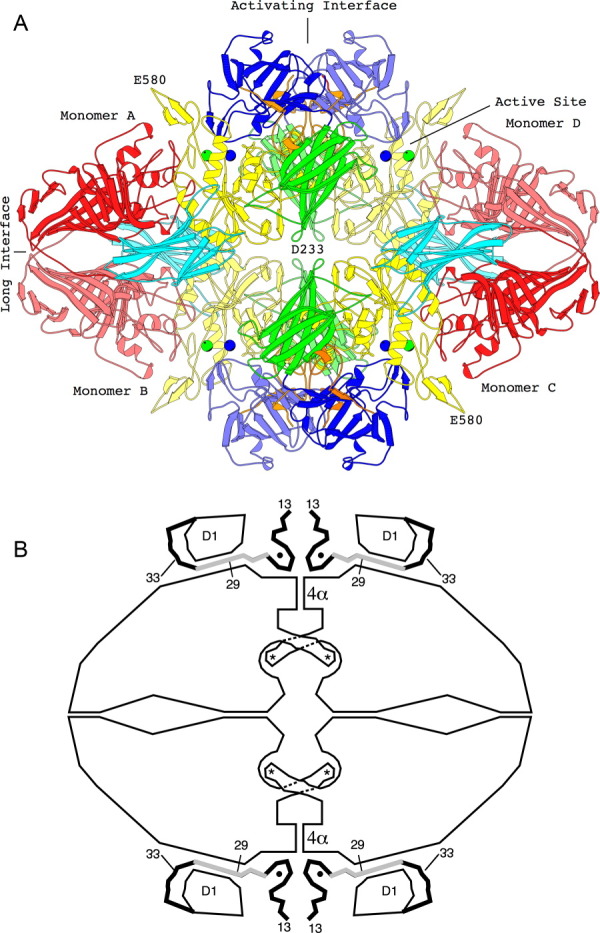
The tetrameric structure of β-galactosidase. (a) The backbone structure of the enzyme. Domain 1, blue; Domain 2, green; Domain 3, yellow; Domain 4, cyan; Domain 5, red. Lighter and darker shading is used to differentiate equivalent domains in different subunits. Metal ions are shown as spheres, Na+, green; Mg++, blue. Interactive views are available in the electronic version of the article (see below). (b) Sketch of the tetramer, aligned as in (a) showing features particularly relevant to α-complementation. The four active sites (each highlighted with an asterisk) are located toward the center of the figure. In each case a loop including residues 272–288 extends from one subunit to complete the active site of a neighboring subunit. The “activation interface” extends vertically through the center of the tetramer. Part of the interface comprises a bundle of four α-helices labeled 4α. Residues 13–50, shown as thick lines, pass through a tunnel between the first domain (labeled D1) and the rest of the protein. The region shaded gray (residues 23–31) is deleted in one of the α-donors. Magnesium ions (small solid circles) bridge between the complementation peptide and the rest of the protein (from Ref. 8). An interactive view is available in the electronic version of the article.
