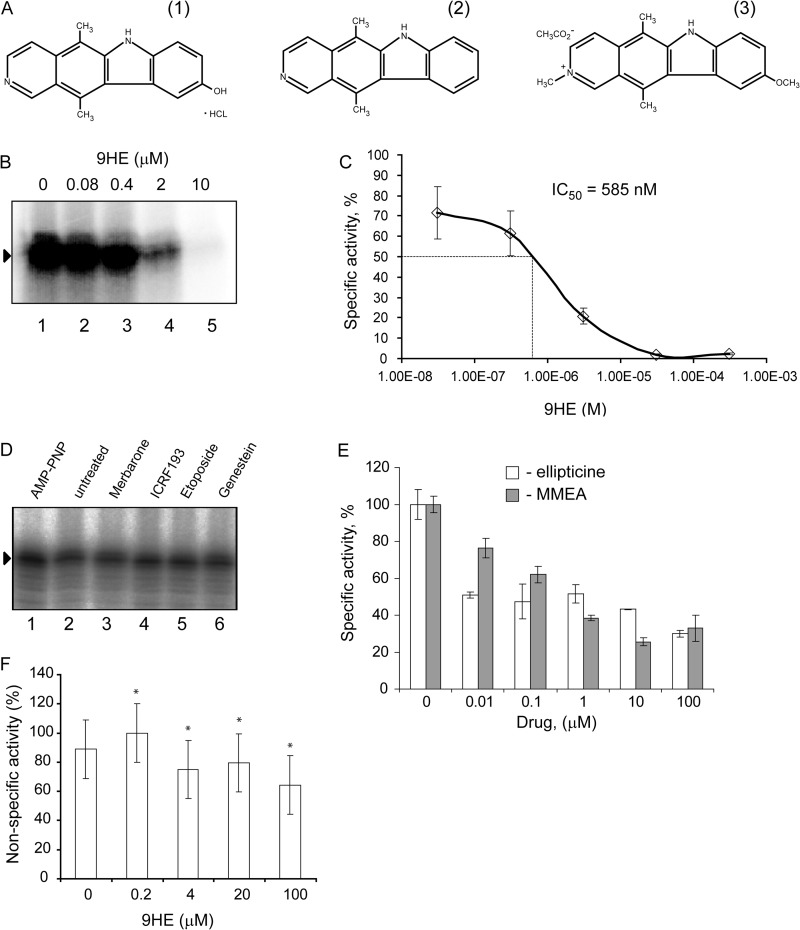
FIGURE 1.
Ellipticines inhibit Pol-I transcription in vitro. A, chemical structures of 9HE (1), ellipticine (2), and 9-methoxy-N2-methylellipticinium acetate (3) are shown. B, specific transcription reactions containing 2 μl of HeLa NE and 200 ng of supercoiled plasmid DNA template were supplemented with various amounts of 9HE as indicated. To ensure an equal loading of the DNA template and NE, the reaction mixture containing all components (excluding drugs) was first prepared, then aliquoted into individual reactions, and then the drug or buffer were added. Transcripts were analyzed by S1 nuclease protection; a representative image is shown. Lane 1 is the control reaction containing no drug. C, the results were quantified with the aid of phosphorimaging. The data are expressed as a percentage of the highest value (set at 100%) and were used for IC50 calculation. The data represent an average from three independent experiments. S.D. is shown. D, specific transcription reactions containing 2 μl of HeLa NE and 200 ng of supercoiled plasmid DNA template were supplemented with different Top2 inhibitors (merbarone, etoposide, and genistein were used at 100 μm final concentration; ICRF-193 at 50 μm, and AMP-PNP at 500 μm). To ensure equal loading of the DNA template and NE, the reaction mixture containing all components (excluding drugs) was first prepared, then aliquoted into individual reactions, and then the drug or buffer were added. Transcripts were analyzed as in B, and representative image is shown. Lane 2 is the control reaction containing no drug. E, specific transcription reactions (as in B) were supplemented with various amounts of ellipticine or 9-methoxy-2-methylellipticinium acetate (MMEA) as indicated. Transcripts were analyzed as in B and quantified as in C. F, nonspecific transcription reactions were supplemented with 1 μl of HeLa nuclear extract and various amounts of 9HE as indicated. The data expressed as a percentage of the highest value (set at 100%) represent an average from three independent experiments. S.D. and statistical significance (*, p < 0.05) are shown. p values have been calculated using one and two-way analysis of variance on R software.