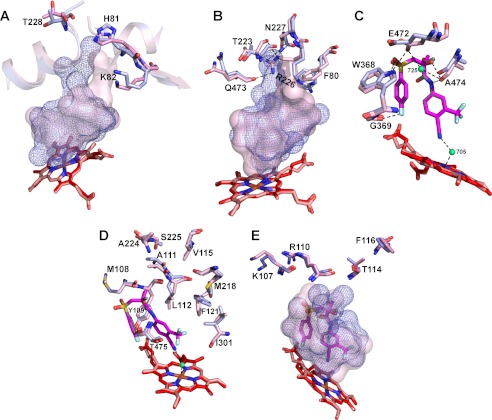
FIGURE 7.
Superimposed views of the active sites in BIC-bound (light magenta) and ligand-free (light blue) CYP46A1 showing the amino acid residues undergoing conformational changes upon BIC binding. The putative sequence of the conformational shifts is also shown. A and B, shown are the movements induced by the initial BIC recognition on the CYP46A1 surface and subsequent BIC sliding inside the substrate access channel. C, shown are key conformational changes in the active site. D, shown are secondary adjustments in the active site. E, shown are secondary adjustments on the CYP46A1 surface. The enclosed volumes of the active site in BIC-bound and ligand-free CYP46A1 are shown as a semitransparent pink surface and blue mesh. Water molecules are shown as green spheres. Dashed black lines indicate hydrogen bonds. The heme group in BIC-bound and ligand-free CYP46A1 are in red and salmon, respectively. The nitrogen, oxygen, fluorine, sulfur, and iron atoms are in blue, red, pale cyan, yellow, and orange, respectively.