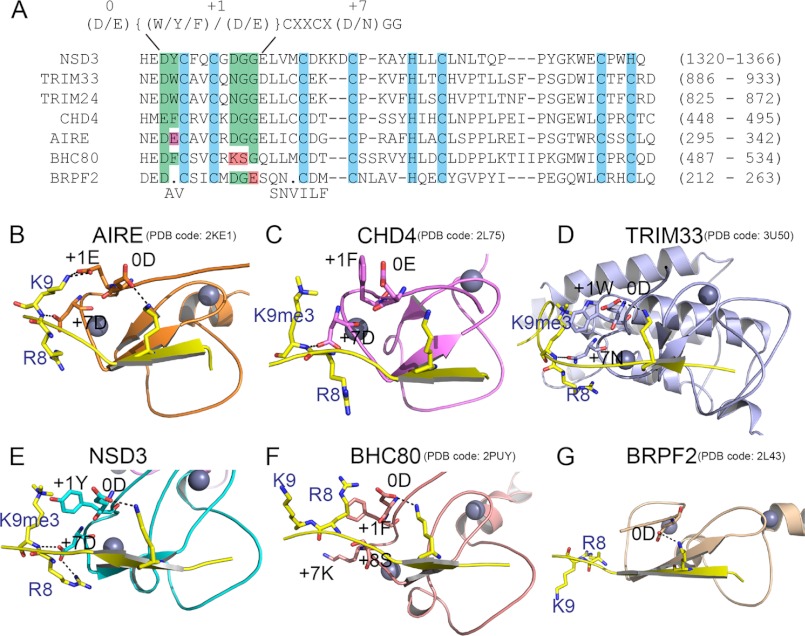
FIGURE 4.
The NTS motif in PHD fingers for H3K4me0 and H3K9 cross-talk. A, sequence alignment of PHD fingers that are known to conduct H3K4me0 and H3K9 cross-talk, including PHD5 of NSD3, PHD1 of AIRE, the PHDs of TRIM33 and TRIM24, and PHD2 of CHD4. The PHD finger of BHC80 and PHD1 of BRPF2, which are not involved in cross-talk, are also included in the alignment. Key residues involved in H3K4me0 and H3K9 recognition are grouped by green background. The conservative NTS motif dedicated to H3K4me0 and H3K9 dual readout is listed at the top of the alignment, with specific positions numbered above. B–G, ribbon representations of the structures of PHD·H3 complexes that are indicated in the alignment in A. The H3 peptides are colored yellow with K4, R8, and K9 or K9me3 side chains shown in stick representation. Key residues of the PHD fingers involved in the recognition of H3 are also shown as sticks.