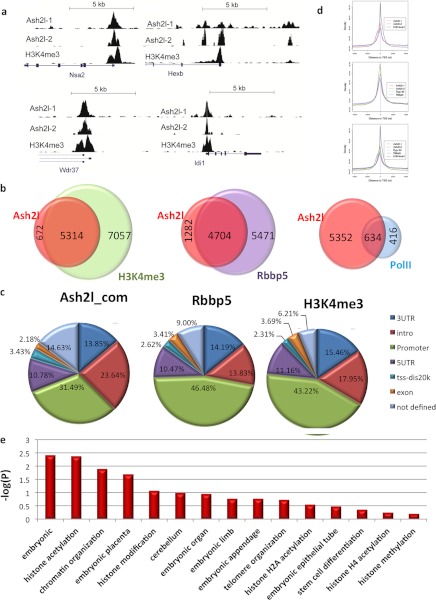
FIGURE 3.
Whole-genome ChIP-seq analysis of Ash2l. a, representative Ash2l binding profiles at its target genes from whole-genome ChIP-seq analysis. The binding patterns of Ash2l on Nsa2, Hexb, Wdr37, and Idi1 were compared with those of H3K4me3. Peak detection was performed with model-based analysis of ChIP-seq (version 1.40) using default settings (p < 10−5). b, a significant portion of ChIP-seq identified Ash2l target genes overlap with those of H3K4me3, Rbbp5, and PolII. c, distribution of Ash2l ChIP-seq binding regions was presented. For comparison, similar analysis was performed for Dpy-30, Rbbp5, and H3K4me3, based on published data sets. Ash2l_com, data from both ChIP-seq experiments. d, binding site distribution of Ash2l, H3K4me3, Dpy-30, and Rbbp5 relative to the transcriptional start sites of RefSeq genes. e, gene ontology analysis of Ash2l-bound genes identified from our whole-genome ChIP-seq experiments. TSS, transcription start site.