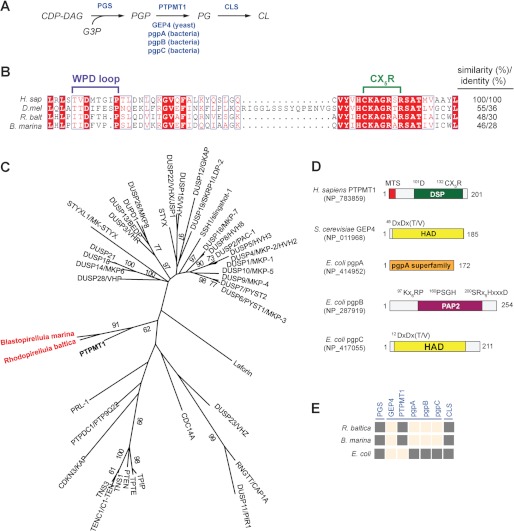
FIGURE 1.
Identification of a putative PTPMT1 ortholog in R. baltica. A, schematic diagram of the CL biosynthesis pathway. Enzymes are highlighted in blue. CDP-DAG, cytidine diphosphate diacylglycerol; G3P, glycerol-3-phosphate. B, active site primary sequence alignment of PTPMT1 orthologs using PROMALS3D and illustrated by ESPript (49, 50). H. sap (H. sapiens), NP_783859; D. mel (D. melanogaster), NP_732901; and R. balt (R. baltica), NP_865112. C, a phylogenetic tree of human and bacterial DSPs constructed using maximum likelihood (ML) method in PHYML. Each branch was tested by 100 bootstrap replicates, and only branches with bootstrap values above 50% were shown. Human PTPMT1 is bolded and bacterial PTPMT1 orthologs are highlighted in red. D, domain architecture of PGP phosphatases. All domains are presented based on analyses using PFAM, CDD, and PROSITE. Catalytic active site sequences are indicated above. MTS, mitochondrial targeting sequence; DSP, dual specificity phosphatase; HAD, haloacid dehalogenase; PAP2, phosphatidic acid phosphatase 2. E, presence and absence matrix for CL de novo synthesis enzymes in Rhodeopirellula. Dark gray squares indicate presence of CL enzyme (column) in target species (row). Peach squares mean no ortholog has been identified, i.e. blast search failed to identify sequences that meet the bidirectional-best-hit criterion (cut-off E-values of 10−10 or less). Sequences of E. coli pgsA, S. cerevisiae GEP4, H. sapiens PTPMT1, E. coli pgpA, pgpB, pgpC, and E. coli cls1 were used as queries for the blast searches.