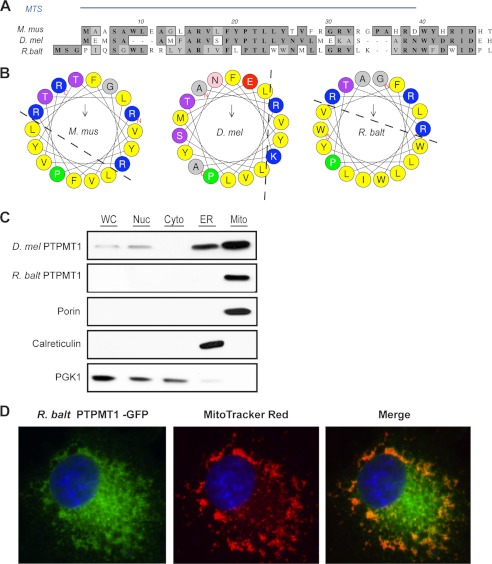
FIGURE 4.
Mitochondrial localization of the Rhodopirellula PTPMT1. A, N-terminal sequences of PTPMT1 orthologs are aligned using PROMALS3D. Amino acid similarities are shaded in light gray and identities in dark gray. The MTS of mouse PTPMT1 has been previously characterized (8) and is indicated by the blue bar. B, N-terminal region of bacterial PTPMT1 forms an amphiphilic helix. 18 amino-terminal residues were plotted on the helical wheel using HeliQuest (51). Basic residues are colored blue, and hydrophobic residues are colored yellow. C, subcellular fractionation of yeast gep4Δ cells expressing fly or bacterial PTPMT1. 30 μg of protein from each fraction were analyzed by SDS-PAGE and immunoblotting. Porin, Calreticulin, and PGK1 serve as marker proteins for mitochondria, ER, and cytosol, respectively. D, bacterial PTPMT1 localizes to the mitochondria of mammalian COS-1 cells. 24 h post-transfection, cells were stained with MitoTracker Red, fixed, and analyzed by immunofluoresence imaging.