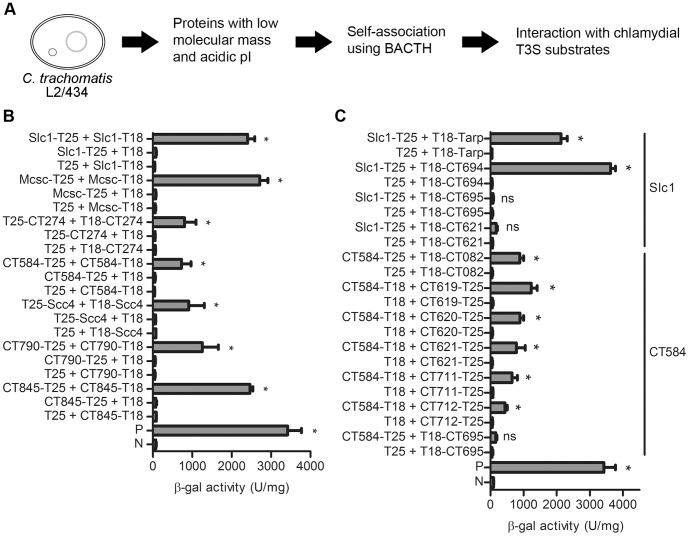
Figure 1. Bacterial two-hybrid screen for novel type III secretion (T3S) chaperone-substrate complexes of C. trachomatis.
(A) The open reading frames within the genome of C. trachomatis L2/434 strain were analyzed for putative proteins with a predicted molecular mass of <30 kDa and isolectric point (pI) of <6 or with a molecular mass of 10–20 kDa and a pI of 6–7. We then selected those showing amino acid similarity to known T3S chaperones or with an unknown function. To determine if the 21 proteins thus identified could form dimers, we analyzed if they self-interacted using the bacterial adenylate cyclase two-hybrid (BACTH) system (B and data not shown). To determine if small and acidic proteins that can self-interact bind C. trachomatis T3S substrates, we systematically analyzed for protein-protein interactions using the BACTH) system (C). (B) and (C) β-galactosidase (β-gal) activity in E. coli BTH101 (cya-99) harboring plasmids encoding the indicated T18 and T25 fusion proteins. P, positive control: E. coli BTH101 expressing fusions of a leucine zipper protein to T18 and T25. N, negative control: E. coli BTH101 expressing T18 and T25. Data are the mean ± SEM from 3 independent experiments. Asterisks denote statistical significant differences relative to the negative control (P<0.05; student’s t-test).