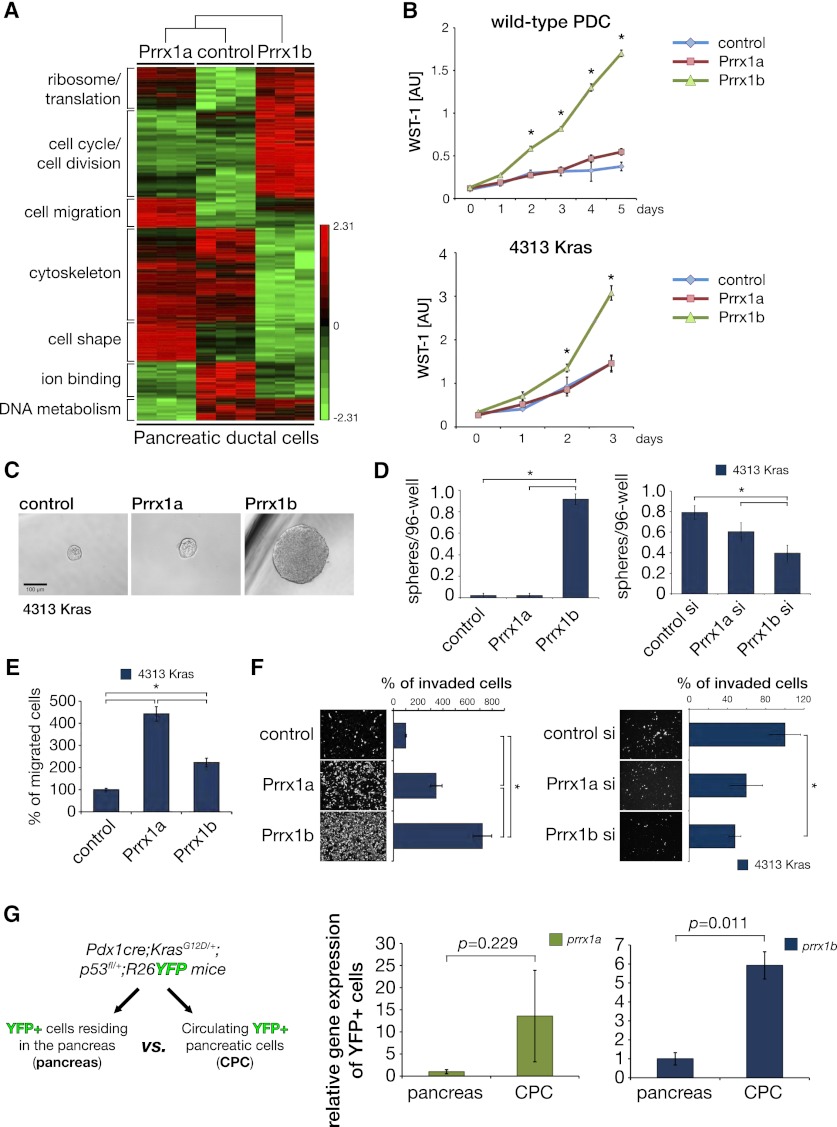
Figure 4.
Prrx1a and Prrx1b display distinct functional properties. (A) Heat map of RNA microarrays of early-passage Prrx1a- and Prrx1b-overexpressing PDCs and control PDCs. GO was performed with DAVID (Huang et al. 2009a,b) and revealed differences in classes of genes related to protein translational machinery, cell cycle/division, cell migration, cytoskeleton, cell shape, ion binding, and DNA metabolism. (B) WST-1 proliferation assay reveals that PDC-Prrx1b cells and 4313-Prrx1b cells have increased proliferation, but not PDC-Prrx1a cells or 4313-Prrx1a cells or control cells. (C,D) Spheroid assay (sphere is defined as diameter >10 cells). 4313-Prrx1b cells form a greater number of spheres than do either 4313-Prrx1a cells or control cells. siRNA Prrx1b knockdown results in a smaller number of spheres. (*) P < 0.05 is statistically significant (Mann-Whitney-Wilcoxon test). (E) Migration assay using 4313 cells overexpressing Prrx1a or Prrx1b, respectively. (F) Boyden chamber invasion assay using 4313 (PanIN). Prrx1a or Prrx1b overexpression in 4313 cells leads to increased invasion, although a greater effect is observed with Prrx1b overexpression. Knockdown of Prrx1b in 4313 cells decreases invasion. (*) P < 0.05 (Mann-Whitney-Wilcoxon test). (G) Prrx1a and Prrx1b mRNA expression (qPCR) in YFP+ CPCs versus YFP+ resident pancreas cells (pancreas) from Pdx1cre;KrasG12D/+;p53fl/+-;Rosa26YFP at a PanIN stage. P-values (paired Student’s t-test) are indicated.