Figure 2.
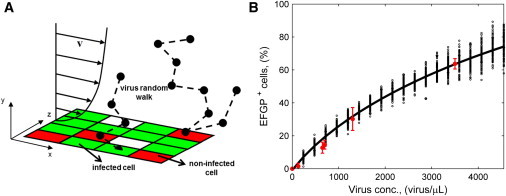
Stochastic model processes and parameter fitting. (A) Graphical representation of the phenomena included in the stochastic model: medium convection with parabolic velocity profile, virus Brownian motion in three-dimensional space, and virus entering a cell with a certain probability when it gets on its surface. Plane x-z represents the bottom surface of the channel where cells are randomly distributed within a regular square grid. (B) Results of model parameter fitting. Experimentally, transduction was performed in a 24-well plate using 200 μL of virus-containing medium for 90 min. Cell concentration was 130 cell/mm2. (Red error bars) Percentage of cells expressing EGFP 24 h after AdV transduction as a function of virus concentration, mean ± standard deviation values obtained in independent experiments, each performed in double or triple. Simulations by the stochastic model reproduce the experimental conditions. In the model, EGFP+ cells are given by cells infected by at least one virus. (Black dots) Model outcome, each dot is obtained from one simulation at the given virus concentration. One-hundred simulations were performed at each condition. (Black line) Simulation mean results.
