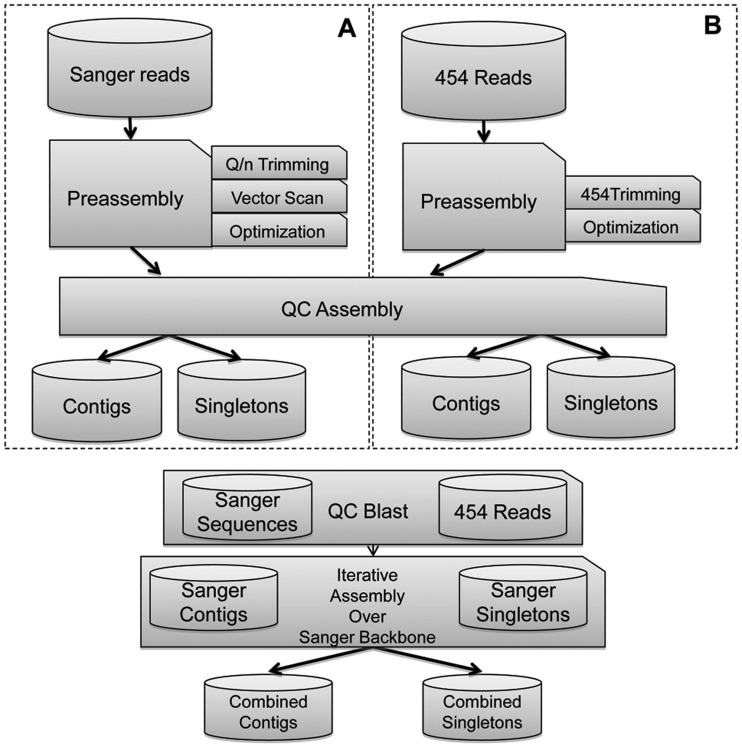
Figure 1.
Workflow followed for the individual assembly of different library/collections and for the assembly of different combinations of library/collections (e.g. assembly of libraries generated from the same tissue). Sections A and B show separate processes for both Sanger and 454 reads assembly. Different assemblies were performed using SeqMan Pro 8.1.1 except when assembling together all the reads generated, which was performed using a two-step assembly process (Newbler 2.6 followed by CAP3) to overcome problems when handling such large amount of data. QC Assembly stands for a quality control step carried out before the actual assembly, where all the reads matching any prokaryotic sequence were removed. QC Blast stands for a quality control step carried out for each assembly consisting of comparing the unigenes generated with themselves using BLAST to guarantee optimal assembly results.