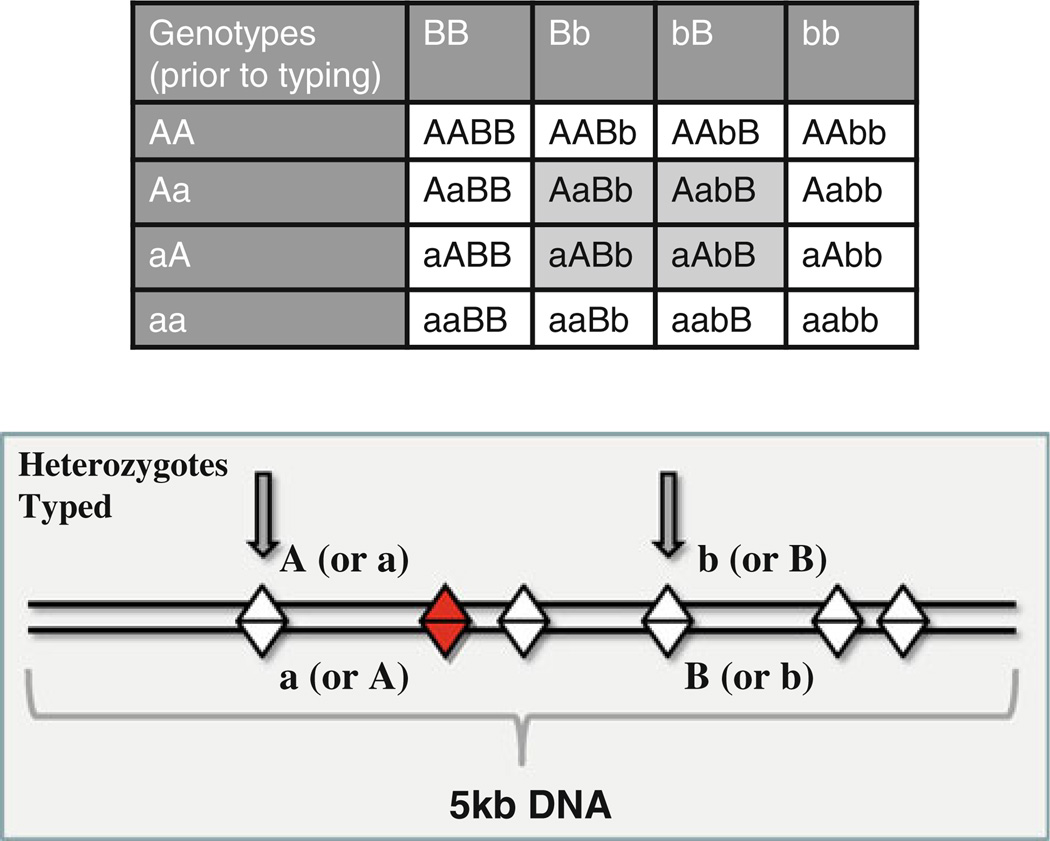
Fig. 2.
Hypothetical SNP chip typing using a two-marker method. Prior to typing, phased diplotypes for the AA, Aa, or aa genotypes and BB, Bb, or bb genotypes could include sixteen possibilities. However, only the four heterozygous combinations (light gray shading) are informative about the phase of the diplotype. The reduction in possible diplotypes when the flanking markers are both heterozygous makes the information gained more powerful. This concept is also depicted in the bottom figure, where straight lines depict a pair of chromosomes, diamonds indicate hypothetical SNP positions along 5 kb of DNA (horizontal lines), the shaded diamond indicates the disease SNP of interest, and arrows indicate the SNPs typed by the chip platform. Thus, when the markers are close enough together so that they are in linkage disequilibrium, two-marker association testing methods have increased ability to find non-typed variants: both SNPs near the disease causing variant combine to reach genome-wide significance