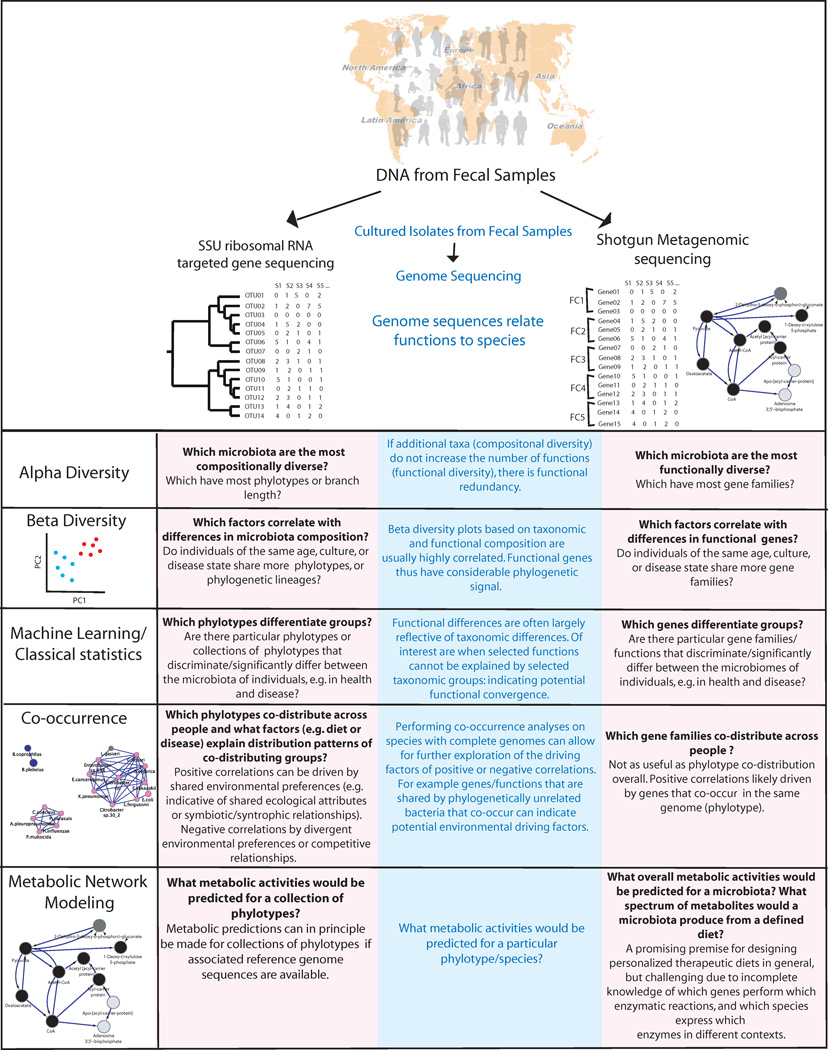
Fig. 2. Tools for understanding compositional and functional diversity of the microbiota, and for generating hypotheses about functionally important genes and how to modulate metabolic phenotypes.
Extracted DNA from fecal samples can be assessed using targeted sequencing of a phylogenetically informative gene (usually SSU rRNA) or random sequencing of all genes. Genome sequences from cultured isolates link these two datasets by indicating which species contain which genes, and therefore functions. Shotgun metagenomic data is thus substantially more useful as the number of reference genomes continues to increase with additional strain sequencing efforts. SSU rRNA gene sequences are usefully related to each other in phylogenetic trees, because related phylotypes (clusters of similar sequences defined by sequence similarity) generally have more similar functional attributes. Functional genes can be binned into functional categories (FC) that are a part of a functional ontology, but those encoding proteins that perform known enzymatic reactions are most usefully related to each other using metabolic networks, because genes that are adjacent in a particular metabolic pathway can produce a phenotype in concert with each other. Compositional and functional diversity patterns can inform each other. They are often highly correlated, but cases where these general correlations do not hold can be biologically or ecologically important. Predicting functions from the species assemblage present still remains an unsolved problem, although the fact that overall genome differences are highly correlated with differences in the SSU rRNA sequence suggests that such predictions may one day be possible. To date, the most powerful studies tend to combine SSU rRNA profiling to determine taxon abundance (the microbiota) with shotgun metagenomic profiling to understand the functions present (the microbiome). Supplementing these studies with mRNA, protein and metabolite level analyses of community samples (and of concurrently obtained host specimens, such as serum and urine) will be crucial so that we can move from in silico predictions of function to direct measurements of expressed community properties.