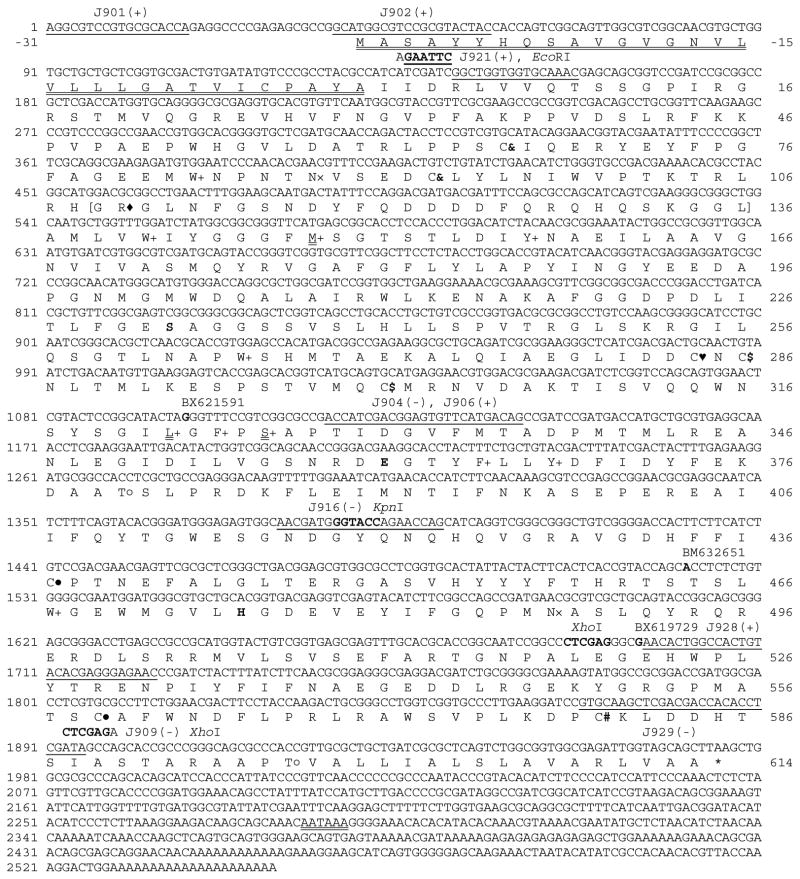
Fig. 1. Nucleotide and deduced amino acid sequences of A. gambiae AChE2.
The last nucleotide of each exon is shaded grey to show the splicing junction. Nucleotides 2203–2484 (CT…AG), missing in cDNA clone BX621591 but retained in clones BM632651 and BX619729, represent a 281 bp cryptic intron of the gene. The starting position of each cDNA is in bold and aligned with its clone name. The polyadenylation signal (AATAAA) is double underlined. Amino acid residues, shown in one-letter abbreviations, are aligned with the second nucleotide of each codon. The predicted signal peptide (−31 to −1) is double underlined, putative N-linked (×) and O-linked GalNAc (○) glycosylation sites are marked, and the stop codon is indicated by “*”. Positions of the Cys residues conserved in all AChEs are marked “& – &, $ – $, and ● – ●” to show disulfide bond connectivity. The unique Cys residue present in many insect AChE1s (Pang, 2006) is replaced by Leu322 (double underlined). Cys284 (♥) is also present in AChEs from D. melanogaster (P07140, Hall and Spierer, 1986) and A. mellifera (AAG43568, Shapira et al., 2001). Cys580 (#) is probably involved in interchain disulfide linkage. The catalytic triad (Ser, His and Glu) is in bold and shaded, and three of the ten conserved hydrophobic residues (+) lining the active site gorge of many AChEs are replaced by M148, L322, and Ser326 (double underlined). The primer binding sites (underlined) and names of restriction enzymes used for cDNA cloning and expression in the baculovirus system are indicated. The cleavage site, located in a hydrophilic region (parenthesized, residues 109–136) of the protein, is indicated by “◆”. The corresponding region (25–30 residues long) is found in some insect AChE2s (Kozaki et al., 2008).