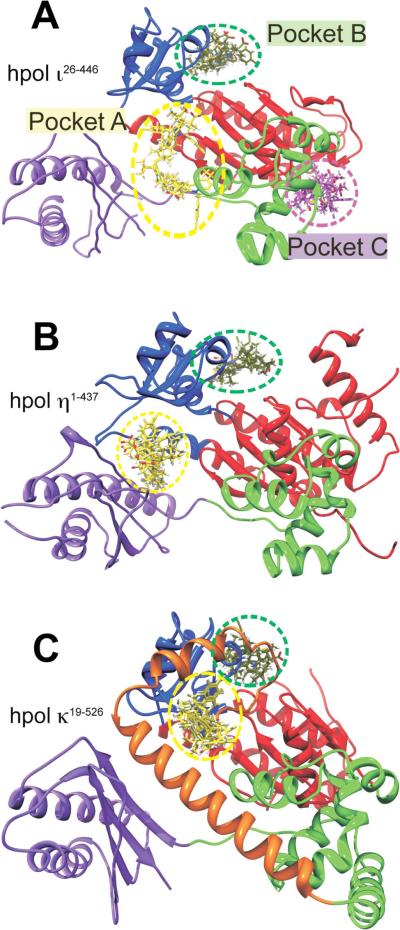
Figure 4.
Docking results for MK886 reveal potential binding sites, including a pocket specific to hpol ι26–446. In silico docking runs were performed as described in Experimental Procedures. A. The crystal structure of hpol ι26–446 (PDB ID 4EBC) is shown as a cartoon representation, with the sub-domains colored as follows: Finger, blue; Thumb, green; Palm, red; little finger (or palm-associated domain), purple. The MK886 binding modes with the highest FF scores from each of the five docking runs are shown as sticks, localized to each of the three binding pockets (A, B and C, indicated by dashed yellow, green and magenta circles respectively). MK886 clusters binding to the pocket A are shown in yellow, pocket B clusters (junction of fingers and palm sub-domains) are in green, while those at pocket C are shown in magenta. Panels B and C show crystal structures of hpol η1–437 (PDB ID 3MR2) and hpol κ19–526 (PDB ID 2OH2), respectively, in the same relative orientation with respect to hpol ι26–446 in panel A, and with the domains colored identically. The `N-clasp' sub-domain in hpol κ19–526 is shown in orange in panel C. Note the absence of any MK886 clusters in hpol η1–437 and hpol κ19–526, in the region corresponding to pocket C of hpol ι26–446. All crystal structure representations shown were generated using the software PyMol (DeLano Scientific, San Carlos, CA).