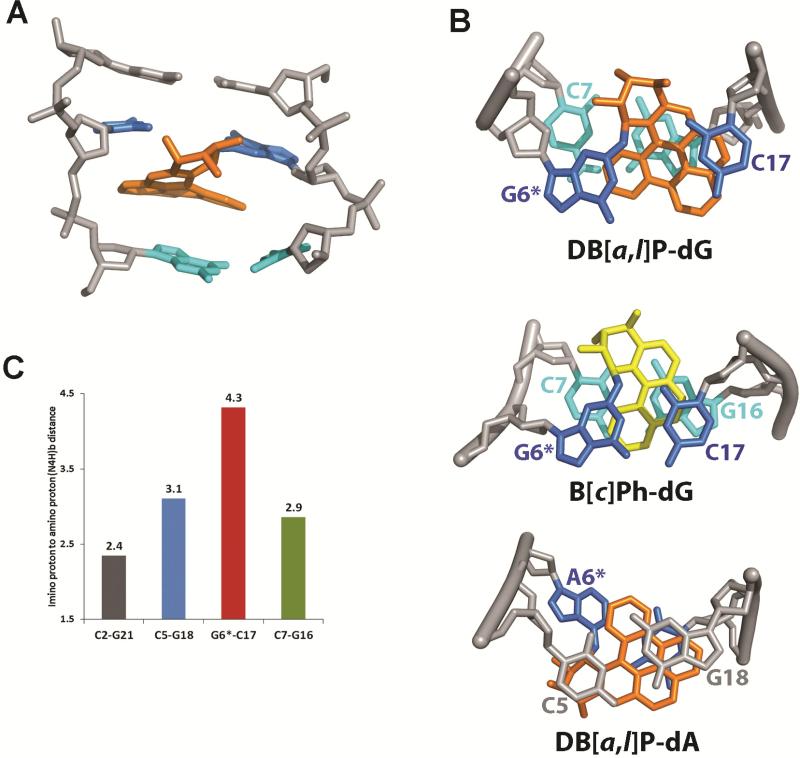
Figure 5.
(A) A DB[a,l]P-dG structure in the 11-mer duplex (Figures 1B,C), selected from the five structures extracted from the restrained MD simulation that best represent the NMR data; the view is from the minor groove of the central trimer. The DB[a,l]P moiety is in orange, the modified guanine and its partner cytosine are in blue, the 3’-side base pair C7:G16 are in cyan, and the rest of the DNA is in light gray. The hydrogen atoms in the DNA duplexes are not displayed for clarity.
(B) Comparison of the stacking patterns of DB[a,l]P-dG, B[c]Ph-dG38 and DB[a,l]P-dA68 adducts in double-stranded oligonucleotides; the views are looking down the helix axis along the 5′→ 3′ direction. In the guanine adducts the DB[a,l]P and B[c]Ph rings are intercalated between G6*:C17 and C7:G16, while for the adenine adduct intercalation is between C5:G18 and G6*:C17. The color code is the same as Figure 5A, except that the B[c]Ph moiety is in yellow. The hydrogen atoms in the DNA duplexes are not displayed for clarity except for the hydrogens of the C7 amino group in DB[a,l]P-dG. A stereoview of the DB[a,l]P-dG adduct is shown in Figure S4 of the Supporting Information. (C) Comparisons of guanine imino to cytosine amino distances (Å) in the NMR distance-refined model (mean values of five structures) for the designated base pairs; the NMR data indicate weakened or absent Watson-Crick pairing at C5:G18, G6*:C17 and C7:G16, while C2:G21 which is more distant from the site of modification has a normal base pair value.