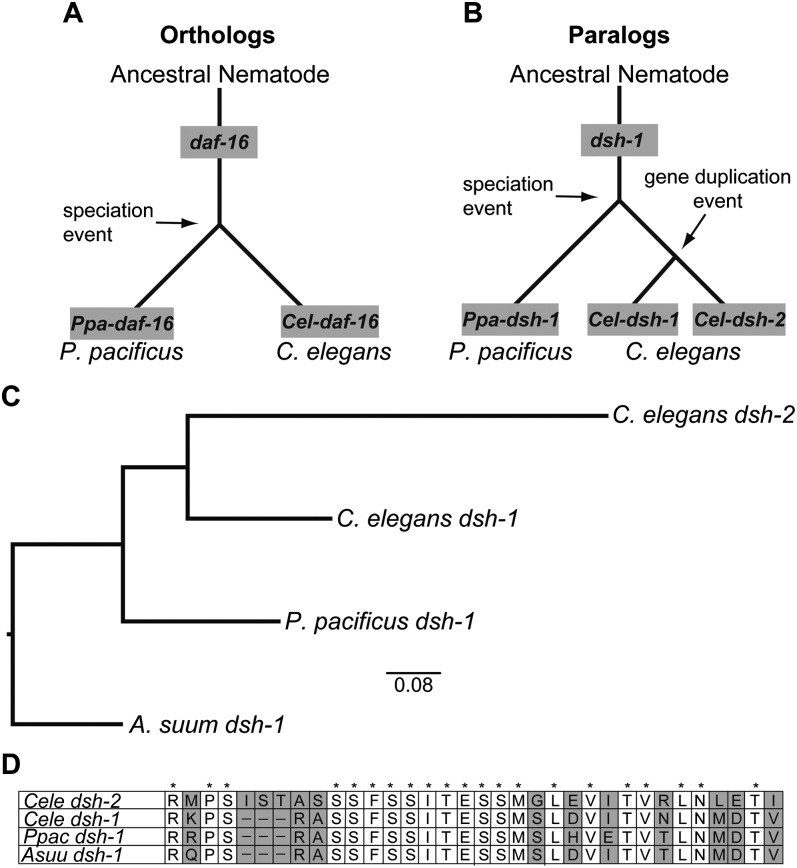
Fig. 4.
Distinction between orthologous and paralogous genes. A) Orthologs are homologous sequences in different species that descend from a common ancestral gene during speciation. The daf-16 gene in an ancestral nematode was conserved in both extant lineages resulting from a speciation event that lead to P. pacificus and C. elegans. Ppa-daf-16 is the conserved daf-16 gene in P. pacificus and Cel-daf-16 is the conserved gene in C. elegans. B) Paralogs are homologous sequences within a species, having arisen by gene duplication or similar event. While Ppa-dsh-1 and Cel-dsh-1 are orthologs in this example, having both been conserved from the same parental dsh-1 copy, Cel-dsh-1 and Cel-dsh-2 are paralogs, having been duplicated within C. elegans. C) Neighbor-joining tree generated from gene comparisons of dsh-1 homologs between A. suum, P. pacificus, and C. elegans, providing evidence that Cel-dsh-1 is the ortholog of Ppa-dsh-1 while Cel-dsh-1 and Cel-dsh-2 are paralogs. The small bar at the bottom center of the tree shows the approximate distance equal to 0.08 nucleotide changes. A. suum was used as the outgroup taxon (see Figure 2). The tree was made from an alignment of the full proteins in MUSCLE and subsequently analyzed using default parameters of the ‘Dnadist’ and ‘Neighbor’ programs from PHYLIP 3.68 software package (Edgar, 2004; Felsenstein, 1989). D) A 34 amino acid window of the protein alignment from which the tree in part C was generated. It shows the sequence conservation of the dsh-1 orthologs and the subsequent divergence of Cel-dsh-2 from the other sequences. Areas of sequence divergence are highlighted in grey while asterisks (*) indicate conserved amino acid identity across all four genes. Hyphens (-) indicate gaps in the alignment, likely the result of insertion/deletion events. A. suum serves as the outgroup taxon.