Abstract
Understanding the principles that govern community assemblages is a central goal of ecology. There is limited experimental evidence in natural settings showing that microbial assembly in communities are influenced by antagonistic interactions. We, therefore, analyzed antagonism among bacterial isolates from a taxonomically related bacterial guild obtained from five sites in sediments from a fresh water system. We hypothesized that if antagonistic interactions acted as a shaping force of the community assembly, then the frequency of resistance to antagonism among bacterial isolates originating from a given site would be higher than the resistance to conspecifics originating from a different assemblage. Antagonism assays were conducted between 78 thermoresistant isolates, of which 72 were Bacillus spp. Sensitive, resistant and antagonistic isolates co-occurred at each site, but the within-site frequency of resistance observed was higher than that observed when assessed across-sites. We found that antagonism results from bacteriocin-like substances aimed at the exclusion of conspecifics. More than 6000 interactions were scored and described by a directed network with hierarchical structure that exhibited properties that resembled a food chain, where the different Bacillus taxonomic groups occupied specific positions. For some tested interacting pairs, the unidirectional interaction could be explained by competition that inhibited growth or completely excluded one of the pair members. This is the first report on the prevalence and specificity of Bacillus interactions in a natural setting and provides evidence for the influence of bacterial antagonist interactions in the assemblage of a taxonomically related guild in local communities.
Keywords: antagonism network, community assemblage, interference interactions, antagonistic interactions, local communities, self-organization
Introduction
Explaining the distribution of microorganisms is a central goal of microbial ecology. A longstanding null hypothesis is ‘Everything is everywhere, the environment selects' (Bass-Becking, 1934; O'Malley, 2007). The appropriateness of this null hypothesis has frequently been questioned, and most of the contentious issues focus on the potential dispersal abilities of microbes. Assembly of organisms in a community is thought to occur through random selection, environmental filtering (abiotic conditions), and biotic interactions. Interest in environmental selection has largely focused on the abiotic environment, for which there are many examples (Hall et al., 2008). In contrast, there has been relatively little investigation on the importance of biotic factors, and in particular on microbial interactions, to explain the distribution of free-living microbes. The organization of a biological community with respect to ecological interactions is referred to as ‘community structure'.
Microorganisms inhabiting a common environment are known to compete for space and resources (Hibbing et al., 2010). In some cases, they excrete chemicals into the environment that are toxic or inhibitory to their competitors. Toxic secretions create antagonist interactions between producers and non-producers, leading to the extinction of the susceptible non-producer in liquid media, but to coexistence in solid structured media (Chao and Levin, 1981; Greig and Travisano, 2008). This coexistence is only achieved by local structure and in the established neighborhood of the contenders (Kerr et al., 2002; Reichenbach et al., 2007; Kim et al., 2008). There are limited reports on the relevance of bacterial interactions in the natural setting, including that of Long and Azam who observed increased antagonism of particle-attached bacteria compared with free-living bacteria from the Pelagic Ocean (Long and Azam, 2001), and that of Mangano et al. (2009), who found that interactions occurring among bacteria isolated from two sponge species may contribute to shaping their bacterial communities. In both studies, the cultured bacterial isolates belong to distant taxons. In fact, community structure has been mainly addressed at the class or phylum level. Regarding intraspecific interactions, Hawlena et al. (2010) showed that distance increased the probability of bacteriocin-mediated inhibition between individuals from two Xenorhabdus species.
Diverse, cultivatable Bacillus spp. lineages have been found to co-occur in the shallow waters along the Churince system in the extremely oligotrophic environment of Cuatro Cienegas Coahuila, Mexico (Cerritos et al., 2010). To gain insight into the assembly of sediment communities and the influence of interfering interactions, we analyzed the interactions between several co-occurring lineages of Bacillus spp. at five physicochemically homogeneous sampling sites and determined the antagonism potential of the isolates against each other within and across the different sampling sites. The null hypothesis of this study assumed that no relation existed between antagonistic interactions and the bacterial assembly at each site, and, therefore, that the frequency of antagonistic interactions among isolates from the five sites would be similar within-site and across-sites. However, our data supported an alternative hypothesis, as there were fewer antagonistic interactions detected within-sites than across-sites, suggesting that antagonistic interactions influenced community assemblage at each site. Furthermore, antagonism and sensitivity could be associated with specific Bacillus taxonomic groups, and network representation of the data revealed a food chain-like structure where the different taxonomic groups occupied a specific place in this hierarchy.
Materials and methods
Site description and sample collection
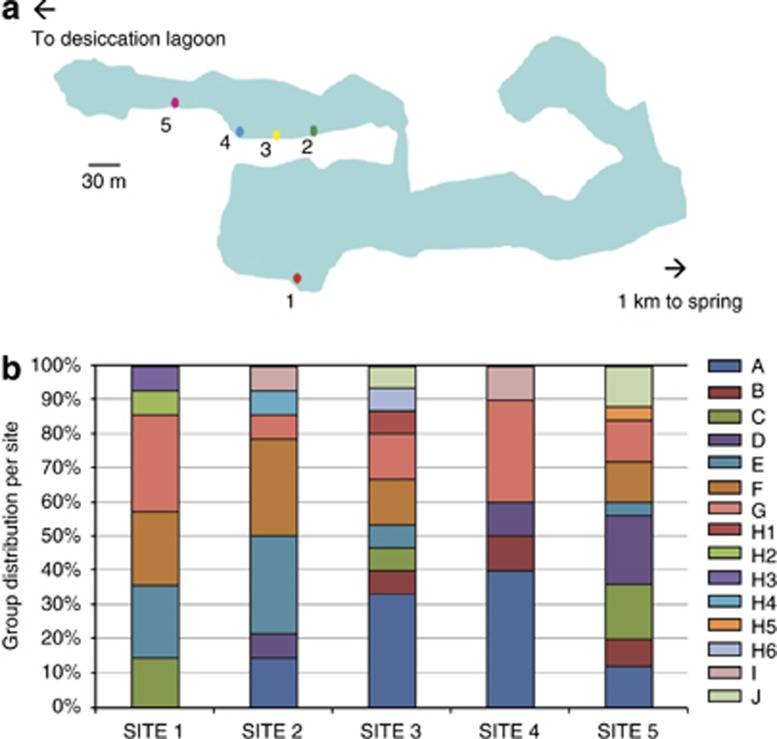
The Churince system consists of three main zones connected by small water causeways, including a spring, an intermediate lagoon and a desiccation lagoon (Johannesson et al., 2004; Carson and Dowling, 2005). Sampling was done only in the intermediate lagoon, which experiences seasonal flooding with minimal flow. Physical chemical measurements revealed seasonal variations but no obvious variations along the sampling points (pH 7.6 to 8; salinity ∼1.7–2.1 ppt, daily temperature fluctuation from 18 to 25 °C in winter and 20 to 35 °C in summer). The sediment description is of gypsum crystals of subangular shape with main grain size distribution between 0.1 and 1 mm. The sampling took place during October of 2007. Five samples were obtained from superficial sediment from various locations along the shore (Figure 1). At each site the top 1–3 mm of the sediment surface were carefully scraped, within a 2 × 2 cm area, and placed in an Eppendorf tube. One milliliter of phosphate-buffered saline buffer was added, and the samples were then incubated at 70 °C for 30 min before plating on marine medium agar plates to enrich for thermoresistant Firmicutes (Cerritos et al., 2008). Colonies of different size, color and morphology were streaked individually a few times until single colonies of a single type were observed. Stocks were prepared in 15% glycerol and stored at −80 °C. Our sample consisted of isolates representative of the diversity observed at each site: 13 isolates from site 1, 14 for site 2, 16 from site 3, 10 from site 4, and 25 from site 5. The distance between each sampling site, from site 1 to 5, was ∼94, 30, 30, 30 and 30 m, respectively.
Figure 1.
Sampling sites in the Churince system and taxonomic distribution of isolates. (a). A schematic map of the Churince system showing the location of the sampling points. (b). The distribution of the isolates belonging to different taxonomic groups is shown as a bar at each site, and the proportion of members belonging to the different groups is shown in different colors. Taxonomic groups A to J were obtained on the basis of the 16S rRNA gene and members in each group have at least 97% identity (see Materials and methods).
Phylogenetic reconstruction
To determine the identity of recovered isolates, we sequenced partial 16S ribosomal (rRNA) sequences. DNA was extracted using the protocol of the QuickGene DNA tissue kit S (Fujifilm Corporation, Minato-Ku, Japan). We amplified the 16S rRNA gene (region V1–V3) by PCR with oligos 16S 27F (5′-AGA GTT TGA TCM TGG CTC AG-3′) and 1492R (5′-TAC GGY TAC CTT GTT ACG ACT T-3′), obtained from Sigma-Aldrich (St Louis, MO, USA). Previous work has shown that this fragment is the most informative for the Bacillus spp. (Goto et al., 2000). The amplicons were sequenced by Sanger (Cinvestav-Langebio, Irapuato, Mexico) using the 16S rRNA gene 27F and 1492R oligonucleotides. The sequences were qualified with Phred (Ewing et al., 1998), and sequences of at least 500 bp were analyzed with the GreenGenes alignment tool (DeSantis et al., 2006), aligned with Muscle (Edgar, 2004), and manual corrections and phylogenetic reconstruction were performed with PhyML (Guindon et al., 2010) using GTR (Generalized time reversible) as a substitution model and a Bootstrap of 1000. Clustering was performed with a 97% threshold using Mothur (Schloss et al., 2009) for the division of the isolates in different taxonomic groups (A to J) (Supplementary Table 1). Six Bacillus spp. sequences did not group within these clades, and these odd sequences were arbitrarily joined as group H for additional analysis (Table 1). GenBank accession numbers of the 16S rRNA gene sequences can be found in Supplementary Table 2.
Table 1. Isolate grouping and the relative frequency of antagonism, sensitivity and resistant phenotypes.
| Groupa | Type of bacteria in the group | No. of isolates in each group | Isolates antagonistic to at least one other isolate | HAb | Sensitive to atleast one other isolate | HSc | fr A | fr HA | fr S | fr HS |
|---|---|---|---|---|---|---|---|---|---|---|
| A | B. pumilus | 14 | 10 | 7 | 8 | 2 | 0.71 | 0.5 | 0.57 | 0.14 |
| B | B. subtilis | 4 | 4 | 4 | 2 | 0 | 1 | 1 | 0.5 | 0 |
| C | B. alcalophilus | 7 | 1 | 0 | 7 | 5 | 0.14 | 0 | 1 | 0.71 |
| D | B. aquimaris | 7 | 5 | 0 | 5 | 4 | 0.71 | 0 | 0.71 | 0.57 |
| E | B. NRRLB14911 | 9 | 6 | 2 | 8 | 7 | 0.66 | 0.22 | 0.88 | 0.77 |
| F | B. cereus | 12 | 6 | 0 | 6 | 0 | 0.5 | 0 | 0.5 | 0 |
| G | B. horikoshii 633 | 13 | 2 | 1 | 11 | 7 | 0.15 | 0.08 | 0.84 | 0.54 |
| H | Diverse Bacillus spp. | 6 | 4 | 1 | 5 | 2 | 0.66 | 0.16 | 0.83 | 0.33 |
| I | Staphylococcus arlettae, S. succinus | 2 | 1 | 1 | 0 | 0 | 0.5 | 0.5 | 0 | 0 |
| J | Diverse Actinobacteria | 4 | 0 | 0 | 4 | 2 | 0 | 0 | 1 | 0.5 |
| Total | 78 | 39 | 16 | 56 | 29 | 0.5 | 0.2 | 0.72 | 0.37 |
Abbreviations: A, antagonistic; HA, highly antagonistic; HS, highly sensitive; S, sensitive.
Most Bacillus groups are related and have at least 97% identity. This classification was based on a 16S rRNA gene alignment, and Mothur was used to generate the phylotype clusters (see Materials and methods). Note that the similarity is not based on the full 16S rRNA gene sequence. Group H includes all divergent Bacillus isolates that have no known strain sequence and that is close enough for comparison (isolates 148, 449a1, 445 and 41b), as well as isolates related to B. indicus (450), B. sp. JAMB-204 (113a), B. megaterium (447) and B. thuringiensis (155a).
HA: isolates that can antagonize 10 or more other isolates were considered highly antagonistic.
HS: isolates that are sensitive to attack from 10 or more other isolates were considered highly sensitive.
Antagonism assays
Assays were performed on solid marine medium using the ‘spot on lawn' method (Burkholder et al., 1966). We used overnight cultures, and every isolate served alternatively as a lawn and as a drop. Square petri dish plates (30 cm) were used to simultaneously test all isolates, and every isolate was spotted on each lawn at least three different times. The plates were incubated for 18 h at 37 °C; inhibition of growth of the isolate on the lawn, observed as a halo around the colony of the spotted isolate, was considered as an antagonist interaction. Assays were carried out to determine if antagonism involved a possible bacteriocin by streaking over the surface of a circular sheet of cellophane covering the plate, that was incubated for 16 h. The cellophane, together with the organisms growing on it, was stripped off, and dilutions of cultures of the test isolates were inoculated as drops on the pretreated medium. After further incubation, the plates were scored for inhibition of growth of the spotted isolates, contrasted with control plates where a non-antagonist isolate was streaked on the cellophane to observe the effect caused by the metabolism of bacterial growth on the medium. Typically, growth of sensitive strains was completely inhibited in medium pretreated by growth of antagonists, and was only minimally affected by growth of non-antagonists (data not shown).
To test for an effect on viability of different interacting isolates and to assay colony morphology in mixes, we plated them on marine agar medium and chose those with phenotypes that could be differentiated in mixes. Selected isolates were grown to stationary phase, and an aliquot frozen at −80 °C in 12% glycerol. Colony forming units per ml were quantified, and the desired number of bacteria were placed, individually or in combinations, in a total volume of 0.5 ml and incubated overnight at room temperature without agitation. Dilutions were then plated out to determine the colony forming units per ml of each of the isolates.
Statistical analysis
Antagonistic interactions were assessed by a nested multifactorial restricted maximum likelihood analysis of variance (JMP 8.1, SAS Institute Inc., Cary, NC, USA), with the producer source site and lawn source site as fixed main effects, and the date and replicate number as random main effects. The producer and lawn strains were included as nested random effects within producer and lawn source sites, respectively. The analysis of variance contained a single interaction effect, which was the producer source site × lawn source site. One planned comparison was performed by comparing antagonism within-sites and across-sites.
Network analysis
We used the degree of vertices, their distribution, assortativity coefficient and motifs to characterize the obtained networks. The degree of a vertex in a graph was considered as the number of edges connected to a vertex. We calculated the input and output degree for each vertex (v), which was denoted as Kin and Kout, respectively. In other words, these values represent the number of isolates inhibiting or inhibited by the isolate. To analyze the network of interactions, we calculated Kin, Kout, and K (total) for each vertex of every network that corresponded to each site, as well as their corresponding probability distributions. Additionally, we examined network assortativity, which indicates the preference of isolates to attach to others that are similar in K. To calculate input, output and degree assortativity coefficients, we used the formula by Newman (Newman, 2002), and to estimate assortativity under other characteristics, we used the formula in Newman (Newman, 2003). Analyses were performed through programming in Octave (http://www.octave.org).
Results
Antagonistic interactions between isolates in local communities are significantly higher in confrontation across-sites
We sampled superficial sediment intermediate lagoon in the Laguna intermedia of the Churince system at five sites, located ∼30 m from each other, with no observed environmental (physical and chemical) differences (Figure 1). Microbes that strongly interact with each other in a microenvironment comprise a local community, and since our interest was centered on testing local bacterial interactions, the size of each of our sediment samples spanned an area of <4 cm2 and was only 1–3 mm thick from the sediment surface. Based on our previous work, Bacillus isolates could be enriched by heating the samples and plating on marine medium (Cerritos et al., 2008), and, therefore, we worked with 78 isolates of cultivated thermoresistant isolates.
The main question in this study was whether isolates co-occurring at each of the different sediment sites, which most likely possessed a history of coexistence, could exhibit differences in antagonism within each site compared with the antagonism they could exhibit when confronted with isolates from a different site. From a 78 × 78 isolate array, 6084 antagonism assays were scored. The most common outcome was resistance (fr=0.91). Only 39 of the strains were antagonistic to any other isolate, and of the other 39 strains able to antagonize one or more isolates, only 516 antagonism events were observed (Figure 2). We analyzed how resistance, sensitivity and antagonistic interactions changed when comparing within-site versus across-site antagonism. We found that there were statistically significant differences in antagonistic interactions within and across-sites, as shown by a planned contrast from a multifactorial analysis of variance (F1,18015=8.33; P=0.0039). Interactions within a site occurred less frequently than interactions across-sites, suggesting that each community assembly had a history of co-existence (Figure 2). The increased frequency of antagonist interactions across-sites suggested that antagonistic traits selects for resistant genotypes, thereby influencing community assembly.
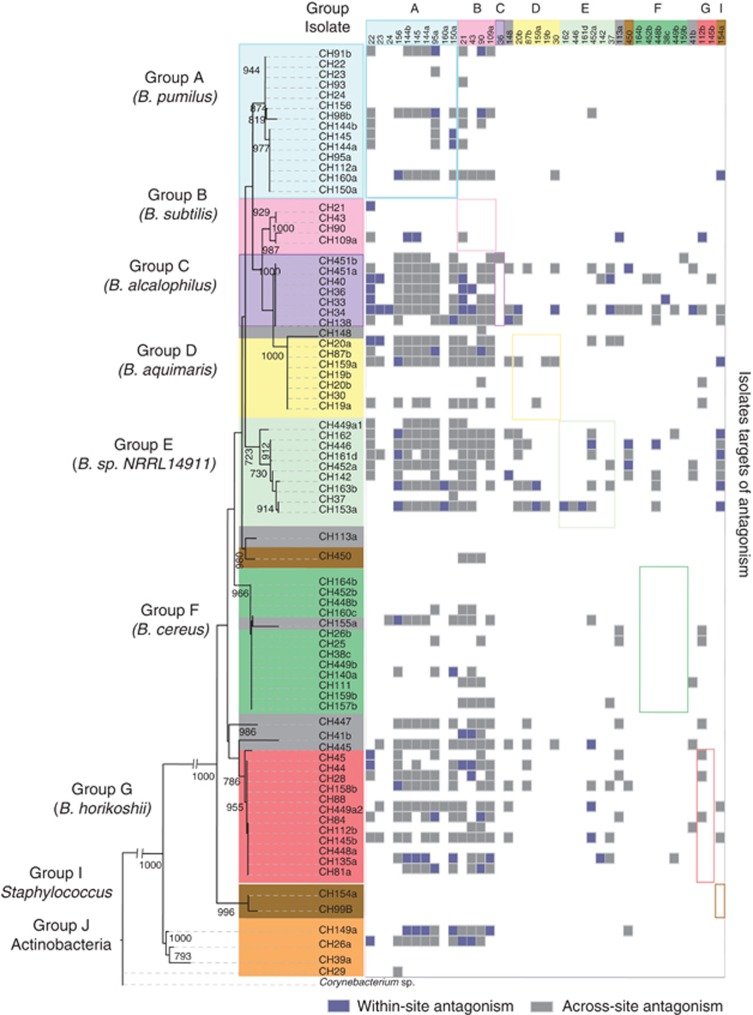
Figure 2.
Taxonomic grouping and antagonist and sensitivity properties of the isolates. On the left a phylogenetic tree of all the strains isolated (78), showing the closest assigned phylotype in each cluster (see Materials and Methods). The top lane of the matrix shows the 39 antagonist isolates; the other 39 isolates did not produce antagonism against any of the 78 isolates tested. Small colored squares in the intersections represent antagonism events where the isolate shown above antagonizes an isolate from the left column. The gray colored squares indicate that the isolates involved have a different site as the source (across-site), and the blue colored squares indicate that both the antagonist and sensitive isolates came from the same site (within-site). Large squares with colored perimeter frame the intersection among members of the same taxonomic group.
Antagonistic interactions of the Bacillus-enriched guild involved at least seven taxonomic groups
A 16S rRNA gene sequence analysis of the isolates revealed that the sample in the study group was formed from 72 Bacillus spp., as well as two Staphylococcus and four Actinobacteria isolates. A phylogenetic tree revealed seven main clades for Bacillus (Figure 2). The distribution of the taxonomic groups varied among-sites (Figure 1). Most of the clades were formed by samples from several sites, and none of the clades were exclusive to any one site. Some cosmopolitan species were closely related to our samples (Table 1). Within the samples, we identified 34 antibiotypes (data not shown) that, together with the differences in colony color and morphology, uncovered a larger phenotypic diversity within the sample than that revealed by the 16S rRNA gene sequence alone, and suggested that the recovered isolates from each site were not clones.
Antagonism is aimed at the exclusion of members of unrelated taxa and, together with resistance, constitutes hidden genetic variability
We observed that antagonistic genotypes were traits of certain taxonomic clades and that the target usually belonged to groups from a different lineage. Lineages A and B exhibited the most antagonistic capacity (ten or more target isolates) (fr=0.42 and 1, respectively) (Table 1), and also exhibited higher resistance than other taxonomic groups. Notably, most antagonism events for the different taxonomic groups occurred outside of the group (fr=0.93) (Supplementary Table 3 and Figure 2). Group C was the most sensitive to Groups A and B (fr=0.34 and 0.28, respectively), followed by Groups D, E and G (Supplementary Table 4). Group F exhibited the highest ability to resist attack from A and B. These data strongly suggested that the consequence of antagonism was mainly the exclusion of members of different Bacillus taxonomic groups. Our data also revealed genetic variability for antagonism. Among genotypes from groups A and B, there was variability in their ability to antagonize or resist attack. For instance, two isolates (CH93 and CH112a) that were non-antagonist were highly resistant, while four others (CH156, CH95a, CH22 and CH150a) were both highly antagonist and highly resistant.
Distribution of the different Bacillus lineages among the sampled sites may explain some of the site's antagonistic properties. For instance, site 1, the most distant of the sampled sites, was the most different in taxonomical composition, lacking members from lineages A and B. As these clades had the most antagonistic isolates, this explained why this site exhibited less antagonism. However, given the antagonist phenotypic variability, the mere isolation of a member of a given lineage from a community does not allow to infer antagonistic capacity or sensitivity. Members of group F, for instance, although present at almost all sampled sites (1, 2, 3, and 5), were non-antagonistic nor exhibited resistance within-site. However, 11 of the 12 isolates from this group were found to be sensitive or antagonistic when confronted across-site (See Supplementary Table 2 for a complete list of the antagonism features of the isolates). We believe that antagonistic and resistance capacity constitutes cryptic genetic variability. That is, the taxonomic diversity observed is a fraction of the potential diversity of the population that is only revealed, both in the antagonist and in the resistant bacteria, upon confrontation.
Network representation of the antagonism relationships reveals properties of a food chain
Network models of complex systems are useful for uncovering patterns that can reveal organizational constraints and functional similarities. In an antagonism network, the ‘social' interactions can be represented in a network topology in which each isolate is depicted as a vertex that can be connected to another vertex. The connecting lines represent antagonism and the number of connections coming out from an individual vertex reveals the antagonism potential of such isolate. In this type of analysis, isolates that are resistant but that display antagonism are placed on top, and isolates that are antagonized are placed below. We observed that all sampled sites had a similar topology that was hierarchical and exhibited directionality (Figure 3a and Supplementary Figure 1). The interactions can be described and quantified in degrees, with Kout and Kin symbolizing antagonist interactions. The Kin=0 degrees when the isolate is resistant, and Kin>0 degrees when the isolate is sensitive. The networks obtained were closer to a network with power-law degree distribution (Figure 3b), while the degree distribution in a random network generated from the isolates (100 000 × sampling in an in silico experiment) always resulted in a normal distribution. This pattern suggests that the observed topology for each site did not occur randomly. Other interesting properties of the network were reflected by the assortativity coefficient, which describes a hierarchical organization where high-degree vertices preferentially inhibit low-degree vertices. In addition, the motifs in the antagonism network exhibited connectivity patterns that recur in a number of different systems (Kashtan et al., 2004). We also noticed a lack of cyclic networks (a sequence of vertices and edges where the first and last vertex are the same), which confirmed the directionality of the antagonism observed, and 817 feed-forward loops (graphs with three vertices, one of which antagonizes another, and both jointly antagonize the last isolate) were identified as well. Events where more than one isolate antagonized the same target are characteristic of other biological networks, such as food chains.
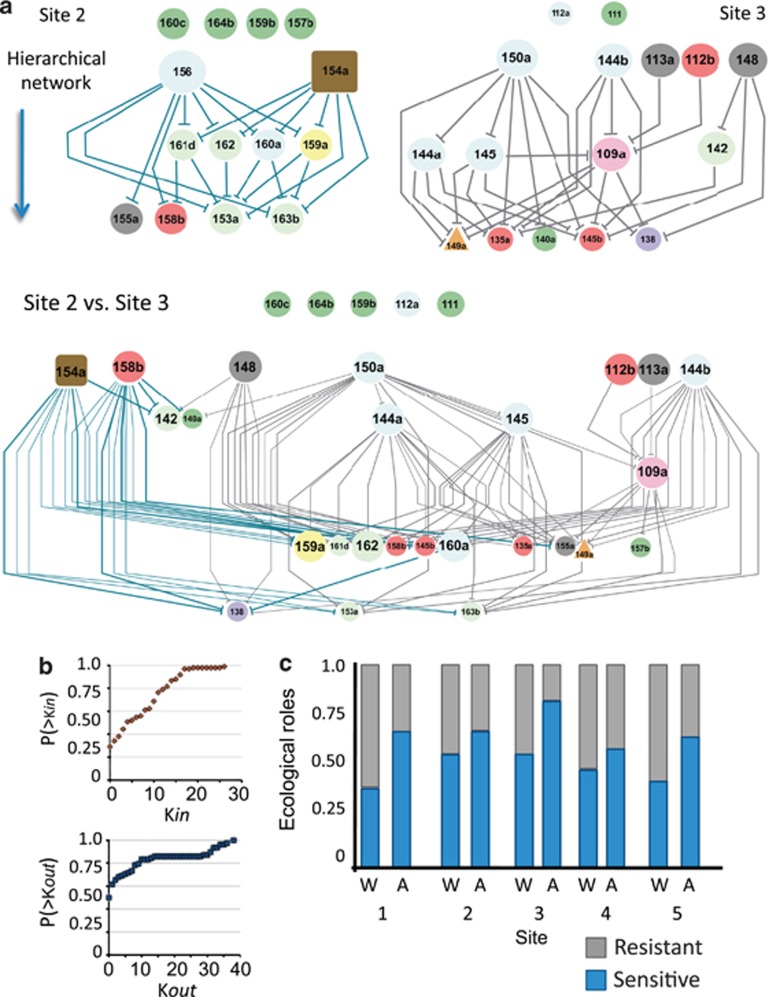
Figure 3.
Network representation of antagonist relationships within-sites and across-sites. (a). Network representation of the within-site and across-site interactions among isolates from sites 2 and 3. Each network represents the hierarchical structure within-site. Each isolate is represented by a vertex, which is colored depending on the taxonomic group to which the isolate belongs (see Figure 2). In addition, the size of the vertex is proportional to the antagonist capability (Kout degree). Vertices of the superior level represent isolates where Kin and Kout=0, followed by Kin=0 and Kout>0, Kin and Kout>0, and finally at the bottom, Kin>0 and Kout=0 isolates. The edges symbolize inhibition interactions between vertices, and the downward direction of these reveals antagonism from vertices in the upper position to vertices at the end of the hierarchy. The bottom network shows the antagonism interactions between isolates from sites 2 and 3. Note that despite the increase in the number of interactions, hierarchy and directionality are maintained. The interaction between isolates with the same inhibitory potential is almost lacking, and, therefore, we found coefficients r<0, which indicate that high-degree vertices preferentially inhibit low-degree vertices. In addition, each assortativity coefficient is statistically significant (greater or less than zero), suggesting that a hierarchical organization exists where the ability of an isolate to inhibit others is not random (see Materials and methods). (b). Analysis of the network topology describes a distribution closer to a power-law degree distribution. We computed the frequency of output and input degrees and their corresponding cumulative degree distributions. Remarkably, half of the vertices had an output degree equal to zero, which means that in the entire network only half of the isolates inhibited another isolate. (c). Higher resistance to antagonism in confrontations among isolates originating from the same site. Graphical representation of the frequency of occurrence of either resistance or sensitivity exhibited by isolates confronted with isolates originating from the same site (within-site) and from a different site (across-sites). These roles are the sum of the degree value for the number of members exhibiting either Kin=0 (resistance) or Kin>0 (sensitivity).
The frequency of antagonism and sensitivity roles within each site is shown in Figure 3c. Notably, the frequency of these roles changed when across-site confrontations were measured. For example, we found that the number of resistant isolates was clearly reduced when across-site antagonism was measured (Figure 3c, gray bars). These findings suggest that antagonism has played a local role in the selection of individuals to co-exist within each site.
Different Bacillus lineages occupy defined positions in the network and differ in their antagonism strategies
We noticed that different Bacillus lineages generally occupied different positions in the antagonism network. B. cereus group (F) members dominated the ‘top' level, and apparently did not interact with the other members, while members of the B. pumilus and B. subtilis, (A and B) were common at the middle and top levels of the hierarchy. The ‘water Bacillus' (C, D, E and G) were almost invariably found at the lowest level, and we noticed that their growth was also less robust (data not shown). Therefore, it seemed as if the architecture of this food chain-like network depended on different lineages at different trophic levels, suggesting a specialization in metabolic functions.
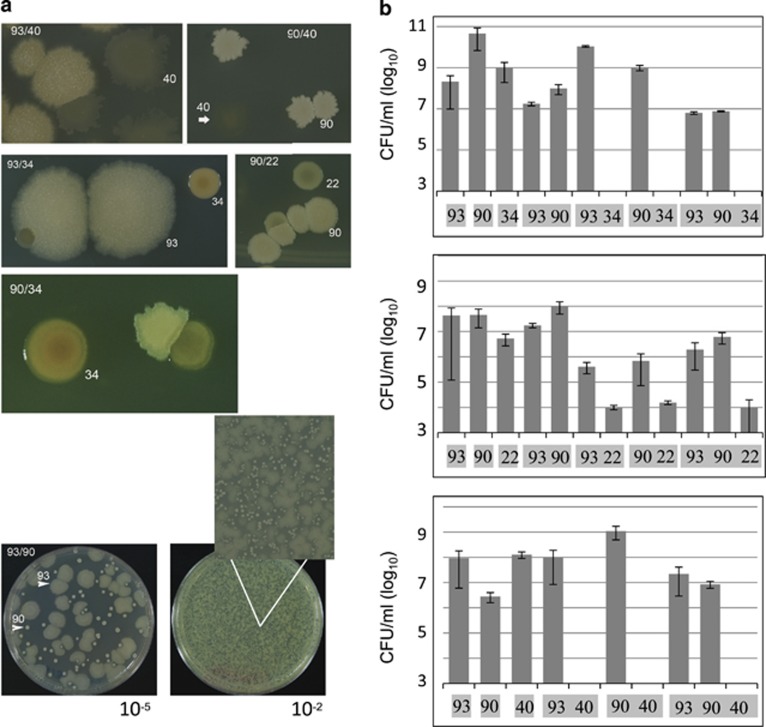
To look closer at the antagonistic interactions uncovered by the network analysis we determined the effect on the viability of isolates tested in pairs and in triads assembled of representative members from the top, medium and bottom levels of the hierarchy. We only worked with those isolates that could be easily identified in particular combinations on the basis of their colony phenotype. Mixes of the different isolates' cultures were prepared and left overnight before plating. Figure 4 shows the results obtained for experiments with isolates CH93 and CH22 (Group A), CH90 (Group B), CH34 and CH40 (Group C). Isolates CH34 and CH40 were only recovered on the plates when added to mixes at 100-fold excess relative to their competitors. However, the colony morphology was drastically affected by the presence of the antagonist isolate (Figure 4a). We observed that isolates from the first and second levels, CH90 and CH93, respectively, competed, but could still co-exist by maintaining distance from each other (Figure 4a). This resulted in a characteristic ‘avoidance pattern' that was noticeable even at high colony density (Figure 4b). The viability of isolates CH34 and CH40 markedly diminished when they were combined with CH93 or CH90 (Figure 4c), or with both simultaneously. Based on these results, we concluded that the antagonistic Bacillus in the Churince sediment excluded sensitive conspecific members, but ‘negotiated' their individual space with resistant members. Finally, antagonism had properties typical of a bacteriocin-mediated inhibition: it acted as a substance that diffused through cellophane, its expression seemed to be regulated (was only produced when the isolates were grown on solid medium and could not be detected in filtered supernatants of isolates grown in liquid medium), and no inhibition was observed when the producer isolate was tested as a target, suggesting there was immunity to their own toxin.
Figure 4.
Antagonism may result in complete elimination or avoidance between isolates. (a). Phenotype of isolates CH34, CH22 and CH40 plated in pairs with either CH93 or CH90 on marine agar medium. When grown with CH93 or CH90, CH34 and CH40 were added at 100–1000 × higher colony forming units per ml to recover them on plates. Interestingly, an ‘avoidance pattern' was clearly observed between CH93 and CH90. Even at lower dilutions, when individual colonies can no longer be observed, this avoidance pattern can be clearly distinguished (see amplification of a small area in the plate). (b). Viability scores of either individual isolates and of each isolate when plated in different combinations. Combination of the isolates resulted in survival of only CH93 and CH90 (at least in the dilutions where isolated colonies could be scored) and reduced the colony forming units per ml of CH22. The viability of isolate CH34 is scored as ‘zero‘, as the typical yellow phenotype was not observed when combined with isolates CH90 or CH93. However, we could only count plates where dilutions resulted in single colonies, and, therefore, we could not determine if the viability count actually decreased to zero.
Discussion
Bacteria in natural environments compete for space and resources. Our study evaluated whether antagonism among a phylogenetically cohesive group can influence their assemblage in a community. Many lineages of Bacillus were found to co-exist within communities in the sediment, and each had differences in their antagonistic and resistance phenotypes. The interactions between isolates can be described by a food chain-like network that shows hierarchy and directionality, where the members at the different levels tend to belong to different taxonomic groups.
Interfering interactions in a Bacillus guild influence their assemblage in sediment communities
Although it is readily assumed that competition for space and resources, as well as the production of substances that can inhibit the growth of other organisms, provides bacteria with an advantage to colonize a niche, there are very few studies to support this assumption in natural environments. Our data on sediment Bacillus spp. suggest that the antagonistic behavior was attenuated within each site, but resistance was lost when across-site antagonism was tested, as expected for a community where the interactions have adjusted to the particular neighborhoods and niches of each taxa. The differences in the intra-site and inter-site frequency observed for each ecological role suggests the occurrence and importance of the antagonism interactions in situ.
Diversity of the isolates, assemblage and competitive exclusion
Relatively little is known about how natural microbial assemblages form and how they are structured. They are often taxonomically complex, and the composition is thought to be based on historical contingency (Martiny et al., 2006). Our analysis of the distribution of isolates from different points in the Churince system showed that numerous bacteria belonging to the Bacillus genus co-occur at the sediment surface (the same seven taxonomic groups have been recovered from the Churince system sediment in at least four new sampling events, data not shown). We assume that these sediment communities draw on the same species pool. As local communities are assembled by migration and depleted by extinction, the patterns of diversity and abundance will depend on the integration and growth success of the migrants influenced by the abiotic environment and the interaction with the established community (Gonzalez and Holt, 2002; Hibbing et al., 2010). Our results suggest that heterogeneity in microbial communities is likely to readily evolve to antagonistic interactions, even over short spatial distances in stream communities, where dispersal is likely to occur with relative frequency. Cryptic genetic variability exists among the species pool. The potential for antagonism, resistance and sensitivity resides in the great diversity of toxins and immunity factors, often carried in phages and plasmids, that can be used to mount strategies to compete for shared resources (reviewed in Kjos et al. (2011)), it is cryptic as is only revealed after chance convergence of bacteria.
It has been suggested that the mechanism underlying the variation in the antagonism effects of toxins like bacteriocins is caused by intraspecific resource competition. Recently Schoustra et al. (2012), showed that antagonism among Pseudomonas aeruginosa strains increased at certain genetic distance. They suggested that targetting a particular group would have a direct effect on the competitors' resource use, and that, therefore, resource use and genetic distance were linked. We suggest that sediment Bacillus compete for resources and that some produce bacteriocins to target resource competitors.
Competition may explain a role in splitting the niche and explain in part the co-occurrence of several lineages of Bacillus within the site, but coexistence of these groups can result from a structured environment that confines the antagonists. Sediment structure, biofilm formation and sporulation would be three mechanisms to shelter isolates from antagonism. Sediment provides a spatial structure that may promote strategies of antagonistic competition by limiting the diffusion of interference toxins and resources (Greig and Travisano, 2008), and may also be necessary and sufficient for the stable coexistence of interacting bacterial species (Kim et al., 2008). It has been shown that a large number of competing species can coexist in a spatially structured habitat (Tilman, 1994).
Antagonism network reveals structure in the interactions among the Bacillus lineages
The antagonism network analysis revealed a hierarchical topology with properties known to be relevant for many biological networks. The power-law distribution results from a large number of vertices with very few connections, while a few vertices have a large number of connections (Barabasi and Bonabeau, 2003). It is known that metabolic networks (Jeong et al., 2000), as well as some but not all food webs (Dunne et al., 2002; Montoya and Sol, 2002), exhibit a power-law degree distribution. This topology is thought to increase network robustness to random vertex or edge deletion but increases sensitivity to a targeted attack (Albert et al., 2000; Dunne et al., 2002). In addition, the analysis of motifs in the antagonism network revealed connectivity patterns that recur in many different systems (Kashtan et al., 2004). A lack of cyclic networks confirmed the directionality of the antagonism observed. This is in contrast to data from Streptomyces (Vetsigian et al., 2011) where reciprocal interactions were commonly observed, particularly among individuals in local communities (individual sand grains). The fact that antagonism is a taxonomic trait of some members of the Bacillus guild explains why the network has direction and hierarchy and its members cannot be randomly connected.
The strong phylogenetic component associated with antagonistic abilities indicates a mode of cryptic niche specialization among Bacillus spp. There seem to be specific niche specialists represented in different clades. Synthesis of antibiotics or toxins has been documented for the Bacillus spp. (Burgess et al., 1991, 1999; Kalinovskaya et al., 2002), and our data is consistent with a bacteriocin-mediated antagonism. However, interference interactions could also result from the synthesis of siderophores, phages, competition for resources, or secretion of end-product metabolites, which could be toxic. Although our results are not a direct measure of the antagonism occurring in situ, if antagonism was only a result of testing under laboratory conditions, then we would not expect differences in the frequency of observed roles when examining isolates within and across-sites. While the interactions we observe are clearly a simplification, modeling of Bacillus spp. interactions reveals important properties that would otherwise be very difficult to disentangle.
It is worth emphasizing that we have only tested the existence of local interactions that reach a few millimeters. This opens the question of whether these interactions can explain the complex spatio-temporal behavior we observed within the sampled regions (in the scale of centimeters), and across sampling sites (in the scale of tens of meters). In this regard, it is important to point out that all the studied bacteria are motile in their local environment, and this presumably enlarges the effective range of the analyzed interactions. On the other hand, it is known that in complex systems with a large number of components that interact locally, global dynamical phenomena, like self-organization can spontaneously emerge. According to Camazine et al. (2003), self-organization is a process in which patterns at the global level of a system emerge solely from numerous interactions among its lower-level components. Moreover, the rules specifying interactions among the system's components are executed using only local information, without reference to the global pattern. Dunstan and Johnson (2005) developed a mathematical model to study the dynamics of marine epibenthic communities, and demonstrated that the patch-scale community patterns of structure and dynamics are emergent, arising from local processes between colonies and species–specific demography. As our system shares many of the characteristics of that studied in Dunstan and Johnson (2005) it would not be surprising that its large-scale spatio-temporal behavior is indeed explained by local interactions like the ones here analyzed. It would be interesting to carry out nested transects at different scales (from millimeter to meter) to determine whether there are patches defined by antagonism and what their limits are.
In conclusion, the observations of ecological interactions among Bacillus spp. from sediment communities are consistent with the hypothesized importance of local antagonistic interactions as an underlying force in the assembly of microbial communities in spatially structured environments. The system global spatio-temporal behavior might be an instance of self-organization, emerging from the local interactions among bacteria.
Acknowledgments
We thank Dr Ma Jesús Puy for sediment description, Francisco Ayala, Mónica Hernández, LP Decena-Segarra, I Corona-Guerrero, VM Delgado-Franyutti, CE, AP Pardo-Arredondo, CE Beltrán-Valdez and MS Torres-Navarro for technical assistance, JA Cisneros and JE Reynoso for help with drawings and photographs, G Bonilla-Rosso for his help performing the Mothur analysis. R-A P-G, VL-R, and LDA were supported by fellowships from CONACYT. This work was financed through a multidisciplinary grant from Cinvestav to GO.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Albert R, Jeong H, Barabasi AL. Error and attack tolerance of complex networks. Nature. 2000;406:378–382. doi: 10.1038/35019019. [DOI] [PubMed] [Google Scholar]
- Barabasi AL, Bonabeau E. Scale-free networks. Sci Am. 2003;288:60–69. doi: 10.1038/scientificamerican0503-60. [DOI] [PubMed] [Google Scholar]
- Bass-Becking LGM. Geobiologie of Inleiding Tot de Milieukunde. W.P. Van Stockum & Zoon: The Hague, the Netherlands; 1934. [Google Scholar]
- Burgess JG, Jordan EM, Bregu M, Mearns-Spragg A, Boyd KG. Microbial antagonism: a neglected avenue of natural products research. J Biotechnol. 1999;70:27–32. doi: 10.1016/s0168-1656(99)00054-1. [DOI] [PubMed] [Google Scholar]
- Burgess JG, Miyashita H, Sudo H, Matsunaga T. Antibiotic production by the marine photosynthetic bacterium Chromatium purpuratum NKPB 031704: localization of activity to the chromatophores. FEMS Microbiol Lett. 1991;84:301–306. doi: 10.1016/0378-1097(91)90373-i. [DOI] [PubMed] [Google Scholar]
- Burkholder PR, Pfister RM, Leitz FH. Production of a pyrrole antibiotic by a marine bacterium. Appl Microbiol. 1966;14:649–653. doi: 10.1128/am.14.4.649-653.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camazine S, Deneubourg JL, Franks NR, Sneyd J, Theraulaz G, Bonabeau E. Self-Organization in Biological Systems. Princeton University Press: Princeton, NJ; 2003. [Google Scholar]
- Carson EW, Dowling TE. Influence of hydrogeographic history and hybridization on the distribution of genetic variation in the pupfishes Cyprinodon atrorus and C. bifasciatus. Mol Ecol. 2005;15:667–679. doi: 10.1111/j.1365-294X.2005.02763.x. [DOI] [PubMed] [Google Scholar]
- Cerritos R, Eguiarte LE, Avitia M, Siefert J, Travisano M, Rodriguez-Verdugo A, et al. Diversity of culturable thermo-resistant aquatic bacteria along an environmental gradient in Cuatro Cienegas, Coahuila, Mexico. Antonie Van Leeuwenhoek. 2010;99:303–318. doi: 10.1007/s10482-010-9490-9. [DOI] [PubMed] [Google Scholar]
- Cerritos R, Vinuesa P, Eguiarte LE, Herrera-Estrella L, Alcaraz-Peraza LD, Arvizu-Gomez JL, et al. Bacillus coahuilensis sp. nov., a moderately halophilic species from a desiccation lagoon in the Cuatro Cienegas Valley in Coahuila, Mexico. Int J Syst Evol Microbiol. 2008;58:919–923. doi: 10.1099/ijs.0.64959-0. [DOI] [PubMed] [Google Scholar]
- Chao L, Levin BR. Structured habitats and the evolution of anticompetitor toxins in bacteria. Proc Natl Acad Sci USA. 1981;78:6324–6328. doi: 10.1073/pnas.78.10.6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunne JA, Williams RJ, Martinez ND. Food-web structure and network theory: The role of connectance and size. Proc Natl Acad Sci USA. 2002;99:12917–12922. doi: 10.1073/pnas.192407699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunstan PK, Johnson CR. Predicting global dynamics from local interactions: individual-based models predict complex features of marine epibenthic communities. Ecol Model. 2005;186:221–233. [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- Gonzalez A, Holt RD. The inflationary effects of environmental fluctuations in source-sink systems. Proc Natl Acad Sci USA. 2002;99:14872–14877. doi: 10.1073/pnas.232589299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto K, Omura T, Hara Y, Sadaie Y. Application of the partial 16S rDNA sequence as an index for rapid identification of species in the genus Bacillus. J Gen Appl Microbiol. 2000;46:1–8. doi: 10.2323/jgam.46.1. [DOI] [PubMed] [Google Scholar]
- Greig D, Travisano M. Density-dependent effects on allelopathic interactions in yeast. Evolution. 2008;62:521–527. doi: 10.1111/j.1558-5646.2007.00292.x. [DOI] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Hall EK, Neuhauser C, Cotner JB. Toward a mechanistic understanding of how natural bacterial communities respond to changes in temperature in aquatic ecosystems. ISME J. 2008;2:471–481. doi: 10.1038/ismej.2008.9. [DOI] [PubMed] [Google Scholar]
- Hawlena H, Bashey F, Lively CM. The evolution of spite: population structure and bacteriocin-mediated antagonism in two natural populations of xenorhabdus bacteria. Evolution. 2010;64:3198–3204. doi: 10.1111/j.1558-5646.2010.01070.x. [DOI] [PubMed] [Google Scholar]
- Hibbing ME, Fuqua C, Parsek MR, Peterson SB. Bacterial competition: surviving and thriving in the microbial jungle. Nat Rev Microbiol. 2010;8:15–25. doi: 10.1038/nrmicro2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong H, Tombor B, Albert R, Oltvai ZN, Barabasi AL. The large-scale organization of metabolic networks. Nature. 2000;407:651–654. doi: 10.1038/35036627. [DOI] [PubMed] [Google Scholar]
- Johannesson KH, Cortes A, Kilroy KC. Reconnaissance isotopic and hydrochemical study of Cuatro Cienegas goundwater, Coahuila, Mexico. J S Am Earth Sci. 2004;17:171–180. [Google Scholar]
- Kalinovskaya NI, Kuznetsova TA, Ivanova EP, Romanenko LA, Voinov VG, Huth F, et al. Characterization of surfactin-like cyclic depsipeptides synthesized by Bacillus pumilus from ascidian Halocynthia aurantium. Mar Biotechnol (NY) 2002;4:179–188. doi: 10.1007/s10126-001-0084-4. [DOI] [PubMed] [Google Scholar]
- Kashtan N, Itzkovitz S, Milo R, Alon U. Topological generalizations of network motifs. Phys Rev E Stat Nonlin Soft Matter Phys. 2004;70:031909. doi: 10.1103/PhysRevE.70.031909. [DOI] [PubMed] [Google Scholar]
- Kerr B, Riley MA, Feldman MW, Bohannan BJ. Local dispersal promotes biodiversity in a real-life game of rock-paper-scissors. Nature. 2002;418:171–174. doi: 10.1038/nature00823. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Boedicker JQ, Choi JW, Ismagilov RF. Defined spatial structure stabilizes a synthetic multispecies bacterial community. Proc Natl Acad Sci USA. 2008;105:18188–18193. doi: 10.1073/pnas.0807935105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjos M, Nes IF, Diep DB. Mechanisms of resistance to bacteriocins targeting the mannose phosphotransferase system. Appl Environ Microbiol. 2011;77:3335–3342. doi: 10.1128/AEM.02602-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long RA, Azam F. Antagonistic interactions among marine pelagic bacteria. Appl Environ Microbiol. 2001;67:4975–4983. doi: 10.1128/AEM.67.11.4975-4983.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangano S, Michaud L, Caruso C, Brilli M, Bruni V, Fani R, et al. Antagonistic interactions between psychrotrophic cultivable bacteria isolated from Antarctic sponges: a preliminary analysis. Res Microbiol. 2009;160:27–37. doi: 10.1016/j.resmic.2008.09.013. [DOI] [PubMed] [Google Scholar]
- Martiny JB, Bohannan BJ, Brown JH, Colwell RK, Fuhrman JA, Green JL, et al. Microbial biogeography: putting microorganisms on the map. Nat Rev Microbiol. 2006;4:102–112. doi: 10.1038/nrmicro1341. [DOI] [PubMed] [Google Scholar]
- Montoya JM, Sol RV. Small world patterns in food webs. J Theor Biol. 2002;214:405–412. doi: 10.1006/jtbi.2001.2460. [DOI] [PubMed] [Google Scholar]
- Newman ME. Assortative mixing in networks. Phys Rev Lett. 2002;89:208701. doi: 10.1103/PhysRevLett.89.208701. [DOI] [PubMed] [Google Scholar]
- Newman ME. Mixing patterns in networks. Phys Rev E Stat Nonlin Soft Matter Phys. 2003;67:026126. doi: 10.1103/PhysRevE.67.026126. [DOI] [PubMed] [Google Scholar]
- O'Malley MA. The nineteenth century roots of ‘everything is everywhere'. Nat Rev Microbiol. 2007;5:647–651. doi: 10.1038/nrmicro1711. [DOI] [PubMed] [Google Scholar]
- Reichenbach T, Mobilia M, Frey E. Mobility promotes and jeopardizes biodiversity in rock-paper-scissors games. Nature. 2007;448:1046–1049. doi: 10.1038/nature06095. [DOI] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoustra SE, Dench J, Dali R, Aaron SD, Kassen R. Antagonistic interactions peak at intermediate genetic distance in clinical and laboratory strains of Pseudomonas aeruginosa. BMC Microbiol. 2012;12:40. doi: 10.1186/1471-2180-12-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilman D. Competition and biodiversity in spatially structured habitats. Ecology. 1994;75:2–16. [Google Scholar]
- Vetsigian K, Jajoo R, Kishony R. Structure and evolution of Streptomyces interaction networks in soil and in silico. PLoS Biol. 2011;9:e1001184. doi: 10.1371/journal.pbio.1001184. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.