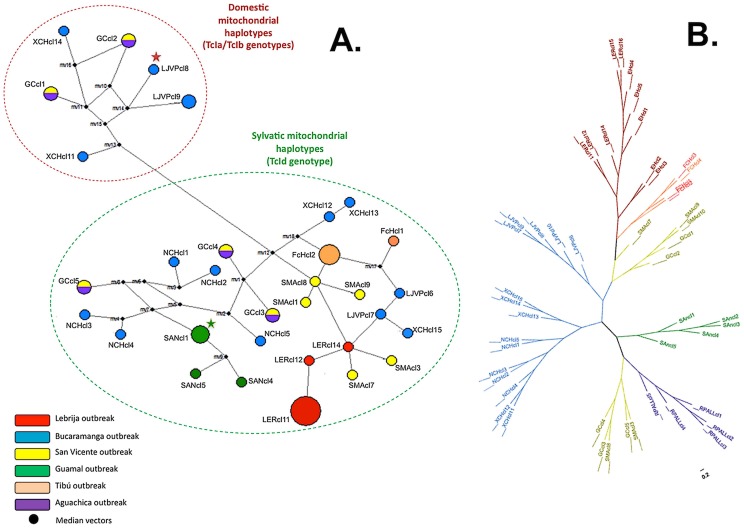
Figure 2. Network haplotype and un-rooted DAS tree of T. cruzi biological closes isolated from the six cases of oral Chagas disease transmission.
A. Median-Joining haplotype network based on MLSTmt of 49 TcI clones isolated from oral outbreaks in Colombia shows the presence of 32 different haplotypes and the discrimination of domestic and sylvatic mitochondrial haplotypes according to the median vectors. Red star indicate the reference sequence of TcI domestic mithocondrial haplotype (EM) and previously typed as TcIa using SL-IR region. Green star indicate the reference sequence of TcI sylvatic mitochondrial haplotype and previously typed as TcId using SL-IR region. There is congruence between MLSTmt and SL-IR genotypes that convey in domestic haplotypes for TcIa/TcIb and sylvatic haplotypes for TcId. The black spots are considered ‘mv’ that can be biologically interpreted as possibly extant unsampled sequences or extinct ancestral sequences. B. Neighbor-Joining genetic distance tree based on MLMT of 49 TcI clones isolated from the oral outbreaks in Colombia shows the clustering of the allelic profiles within each outbreak. A significant number of allelic multilocus genotypes can be considered within each cluster. The topology of the un-rooted tree demonstrates the allelic relatedness among the clones isolated within the same oral outbreak with the presence of some outliers.