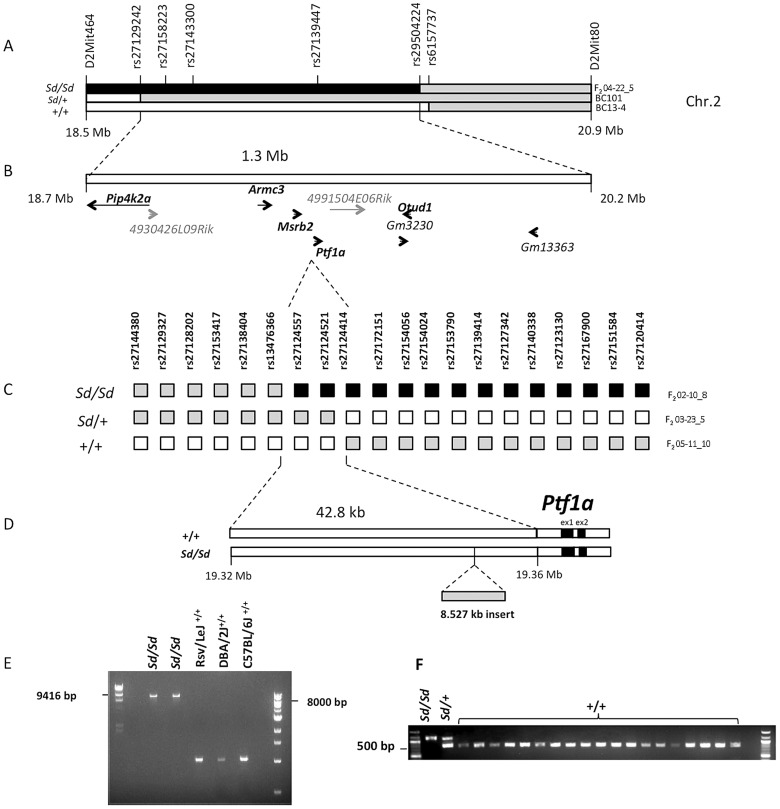
Figure 1. Fine mapping and identification of the Sd mutation.
(A) Meiotic mapping on mouse Chr.2 on 174 backcross (BC) and 120 F2 intercross mice refines the Sd locus to a 1.3 Mb interval. The critical recombinants in 2 BC and 1 F2 mice are shown. The phenotype of each mouse (Sd/Sd, Sd/+ and +/+) is indicated on the left. The genotypes are indicated by the color of each haplotype: black = RSV/RSV genotype; gray = RSV/B6 genotype; white = B6/B6 genotype. The location of genotyped markers is indicated above the haplotypes. (B) The initial mapping localized the Sd mutation to a 1.3 Mb interval on Chr. 2 between rs27129242 and rs29504224. This region contains 9 genes, of which five have mammalian orthologs (shown bold black). Predicted genes without mammalian orthologs are indicated in gray. (C) High resolution mapping in 1203 F2 mice. The critical recombinants in 3 F2 mice are shown. The phenotype and genotypes are indicated as in (A). (D) Localization of the Sd mutation to a 42.8 kb interval between rs13476366 and rs27124414, 12.2 kb upstream of the Ptf1a start site and identification of an insertion of 8.5 kb in the Sd/Sd mouse. (E) Long-range PCR identifies the insertion in 2 different mice Sd/Sd mutants but not in the RSV/LeJ strain and the two counterstrains used in the mapping study (C57BL/6J and DBA/2J). (F) Genotyping identified the insertion in Sd/Sd, and Sd/+ samples but not in any additional mouse strain tested, including 4 wild-derived and 15 other laboratory inbred strains (listed in methods).