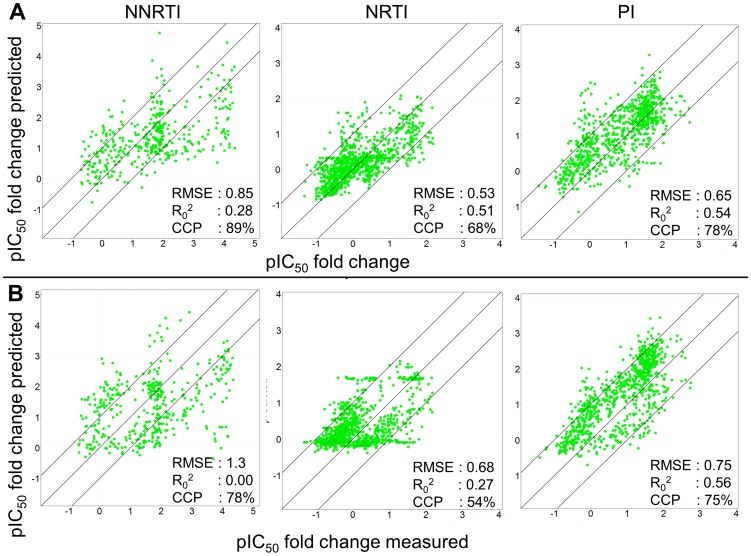
Figure 4. Performance of PCM based models compared with sequence based models for the 150 most difficult sequences as published by Van der Borght et al.
The PCM models (A) perform better as they have a lower prediction error for each drug class (0.53 log units versus 0.68 log units for the NRTIs; 0.65 log units versus 0.75 log units for the PIs, and 0.85 log units versus 1.3 log units for the NNRTIs) than the sequence based models (B). Clearly the NNRTIs are most difficult to predict. Note that these sequences contain a large fraction of mixture sequences, which were not present in the PCM training set but were present in the sequence only training set. In addition, the PCM models also reach a higher CCP compared to the sequence only models.