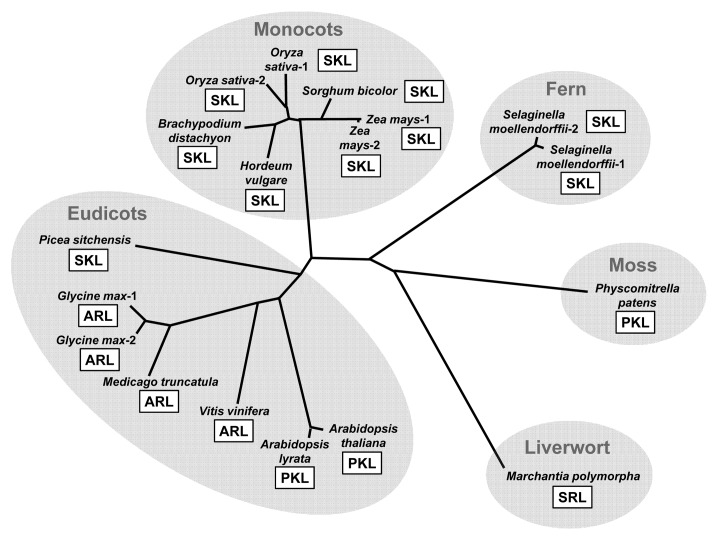
Figure 2. Phylogenetic relationship of the plant KAPA synthases. The amino acid residues of their C-terminal peroxisomal targeting signals (PTS1) are also shown with open squares. The full-length amino acid sequences of the plant KAPA synthases were aligned using Clustal W program (version 2.1) and the unrooted phylogenetic tree was constructed using the neighbor-joining method. The GenBank accession numbers for the sequences retrieved are as follows: Arabidopsis thaliana, NP_974731.1; Arabidopsis lyrata, XP_002871105.1; Oryza sativa-1, BAD87813.1; Oryza sativa-2, NP_001065381.1; Hordeum vulgare, BAK03504.1; Brachypodium distachyon, XP_003574335.1; Sorgham bicolor, XP_002467492.1; Zea mays-1, ACG35792.1; Zea mays-2, ACG35881.1; Selaginella moellendorffii-1, XP_002969752.1; Selaginella moellendorffii-2, XP_002981364.1; Physcomitrella patens, XP_001769874.1; Picea sitchensis, ABR18106.1; Vinis vinifera, XP_002268950.1; Medicago truncatula, XP_003598166.1; Glycine max-1, XP_003527547.1; Glycine max-2, XP_003522881.1. The peptide sequence of the putative Marchantia polymorpha KAPA synthase was confirmed by PCR amplification and sequencing of its cDNA based on the information from the Marchantia expression sequence tag database (S. Yamaoka, unpublished data).

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.