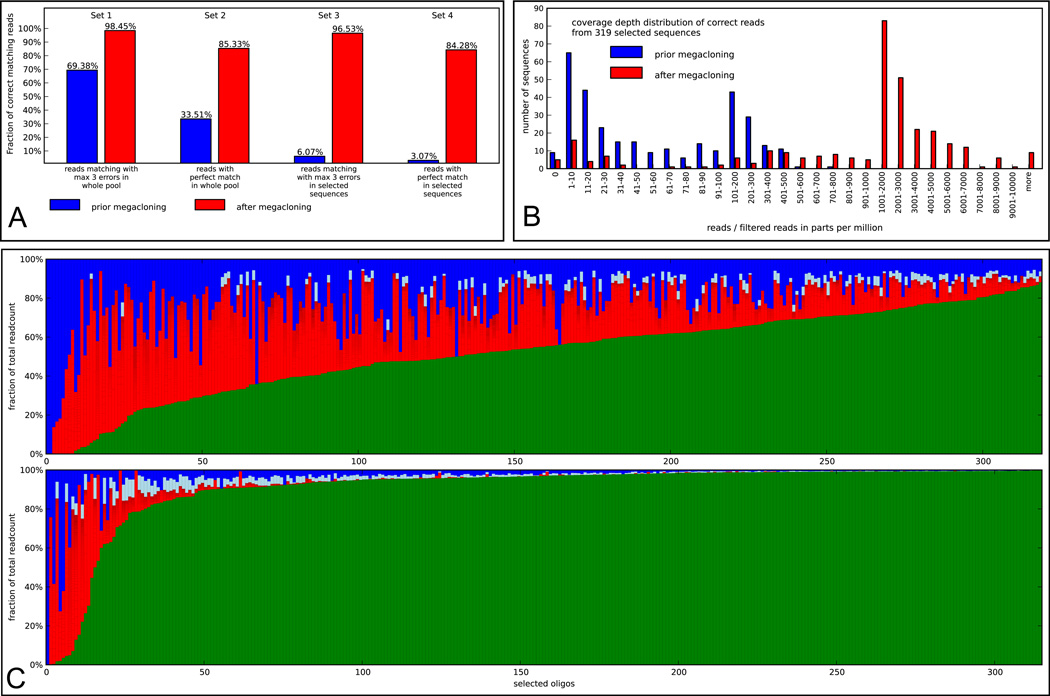
Figure 2.
(A) Comparison of the initial microarray oligonucleotide pool (blue) and the pool enriched with the Megacloner technology (red) based on the results of the Illumina GAII runs. The bars in Set 1 represent the fraction of reads that could be mapped allowing up to three errors, bars in Set 2 show the fractions of perfectly matching reads to the sequence set of the initial pool (3918 sequences). Differences between the blue and the red bar in Set 2 represent the enrichment of correct sequences by Megacloning. The bars in Set 3 and Set 4 show the fractions of reads mapping to sequences from the selected pool (319 sequences). Differences between blue and red bars in Set 3 show the enrichment of selected 319 sequences prior vs. after Megacloning, blue and red bars in Set 4 represent the enrichment of sequences which are in the set of 319 selected sequences and which are correct. (B) Histogram of read counts in the Illumina GAII data of the initial pool (blue) and the enriched Megacloned sample (red). Only reads mapping without errors to one of the 319 selected target sequences have been taken into account. To compare the two NGS runs on the basis of read counts the numbers have been converted into parts-per-million-units (ppm) taking the number of filtered reads as basis. (C) Composition of reads from the Illumina GAII data including 319 selected sequences in the initial pool (top) and the enriched pool (bottom). The oligonucleotides are sorted by the fraction of correct reads. Green: correct reads, red: error prone reads (compartments in the red bars represent single sequences with a readcount of 0.1% or more of total reads for the particular sequence). Light blue: sum of non-unique error prone reads where each sequence represents less than 0.1% of total reads for the particular sequence. Blue: unique reads. In the Illumina GAII dataset from the enriched sample just 315 out of 319 selected sequences could be detected.