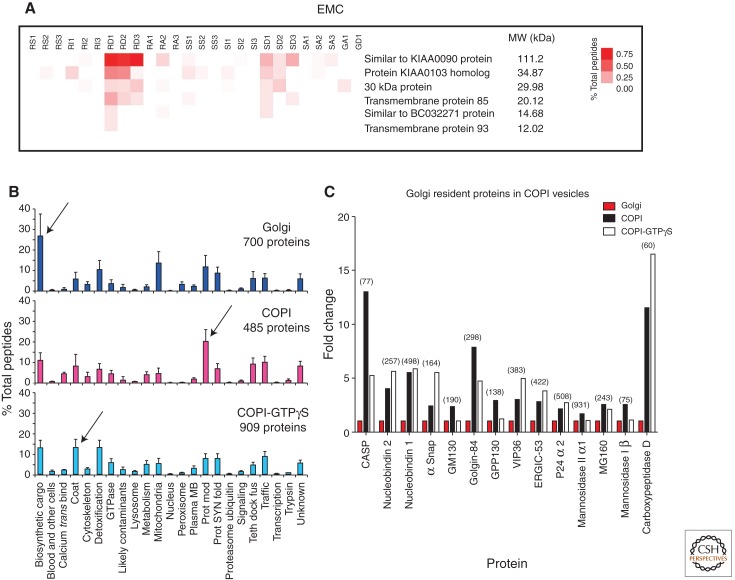
Figure 5.
Proteomics discoveries within the ER and Golgi. (A) Rat homologs of the yeast EMC. Rough and smooth microsomes were subfractionated as described in Gilchrist et al. by salt wash (RS1–3 and SS1–3; for rough and smooth microsomes, respectively), followed by solubilization by the detergent Triton-X 114. The integral membrane proteins partition into the detergent phase (RD1–3 and SD1–3). All six proteins, considered to be homologs of the yeast EMC proteins, are at highest abundance (% total peptides) in the RD. Hence, the EMC proteins are concluded to be integral membrane proteins of the rough endoplasmic reticulum in rat liver. (B) The proteins detected in the Golgi, COPI, and COPI-GTPγS fractions, as organized by functional categories. Arrows indicate functional categories that were most abundant in each fraction (N = 3, means and SD shown). (Data from supplement of Gilchrist et al. 2006.) (C) Golgi resident proteins are enriched in COPI vesicles. When the proteins that increase in COPI vesicles versus the Golgi fraction were interrogated with the Gilchrist et al. (2006) resource, Golgi resident proteins were seen to be more abundant in the two COPI vesicle fractions (total number of peptides identified for each protein indicated in parentheses).