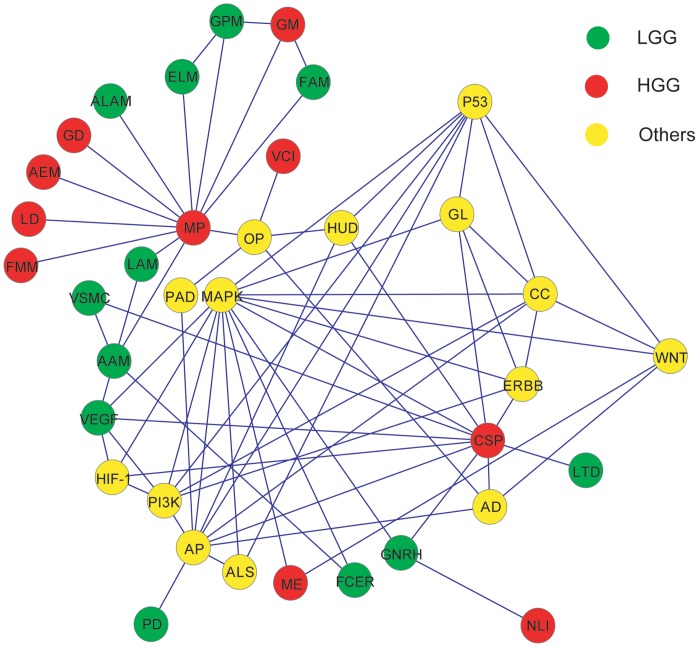
Figure 1. A network of pathways related to LGG, HGG, and others.
The abbreviations for KEGG pathways used in this analysis are listed as follows. AAM: Arachidonic acid metabolism; LAM: Linoleic acid metabolism; ALAM: alpha-Linolenic acid metabolism; ELM: Ether lipid metabolism; GPM: Glycerophospholipid metabolism; PD: Prion diseases; GNRH: GnRH signaling pathway; LTD: Long-term depression; VSMC: Vascular smooth muscle contraction; VEGF: VEGF signaling pathway; FCER: Fc epsilon RI signaling pathway; FAM: Fatty acid metabolism; MP: Metabolic pathways; NLI: Neuroactive ligand-receptor interaction; CSP: Calcium signaling pathway; ME: Melanogenesis; FMM: Fructose and mannose metabolism; LD: Lysine degradation; AEM: Androgen and estrogen metabolism; GM: Glycerolipid metabolism; GD: Glycosaminoglycan degradation; VCI: Vibrio cholerae infection; AD: Alzheimer's disease; PAD: Parkinson's disease; ALS: Amyotrophic lateral sclerosis; HUD: Huntington's disease; PI3K: PI3K-Akt signaling pathway; HIF-1: HIF-1 signaling pathway; MAPK: MAPK signaling pathway; CC: Cell cycle; P53: p53 signaling pathway; AP: Apoptosis; GL: Glioma; ERBB: ErbB signaling pathway; WNT: Wnt signaling pathway; and OP: Oxidative phosphorylation.