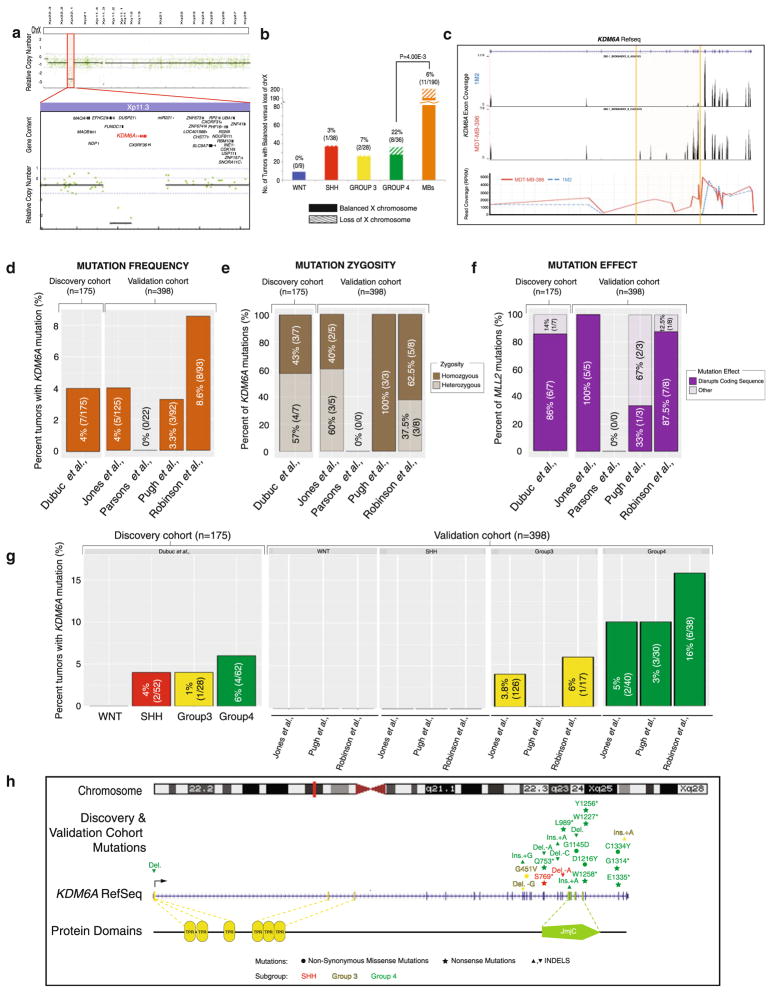
Fig. 2.
Identification of focal deletions of KDM6A, and mutations that are enriched in Group 4 tumors. a Analysis of medulloblastoma copy number aberrations reveals one (0.5 %, 1/203) tumor with a homozygous deletion spanning KDM6A. b Subgroup-specific distribution of X-chromosome loss across 190 primary medulloblastomas studied by Affymetrix 500 K SNP arrays demonstrates a significant (P = 4.00E−3) enrichment of this cytogenetic event in Group 4 medulloblastomas (22 %, 8/36). c Targeted exon-capture and sequencing identifies a medulloblastoma (0.6 %, 1/175, 1M2) with a KDM6A intragenic deletion spanning exons 5 through 13. d Frequency and percent of KDM6A mutations across a discovery cohort of 175 primary tumors (left) and a validation cohort of 398 tumors (right) reveals mutations in 4–8 % of tumors. e Allele frequency inference of KDM6A mutation zygosity highlights the abundance of homozygous mutations across both cohorts. f Percent distribution of KDM6A mutations which disrupt the coding sequence (frameshift or nonsense mutations) versus non-disruptive nucleotide changes (missense and in-frame INDELs) accents the frequent disruptive nature of KDM6A mutations across the discovery and validation cohorts. g Subgroup-association of KDM6A mutations demonstrates predominant targeting of Group 4 medulloblastomas. h Mutational landscape of KDM6A across 573 tumors (combined discovery and validation cohort) calls attention to the 3′ localized, damaging and Group 4-enriched nature of KDM6A single nucleotide variants