Abstract
The structural basis for allosteric regulation of phenylalanine hydroxylase (PAH), whose dysfunction causes phenylketonuria (PKU), is poorly understood. A new morpheein model for PAH allostery is proposed to consist of a dissociative equilibrium between two architecturally different tetramers whose interconversion requires a ~90° rotation between the PAH catalytic and regulatory domains, the latter of which contains an ACT domain. This unprecedented model is supported by in vitro data on purified full length rat and human PAH. The conformational change is both predicted to and shown to render the tetramers chromatographically separable using ion exchange methods. One novel aspect of the activated tetramer model is an allosteric phenylalanine binding site at the inter-subunit interface of ACT domains. Amino acid ligand-stabilized ACT domain dimerization follows the multimerization and ligand binding behavior of ACT domains present in other proteins in the PDB. Spectroscopic, chromatographic, and electrophoretic methods demonstrate a PAH equilibrium consisting of two architecturally distinct tetramers as well as dimers. We postulate that PKU-associated mutations may shift the PAH quaternary structure equilibrium in favor of the low activity assemblies. Pharmacological chaperones that stabilize the ACT:ACT interface can potentially provide PKU patients with a novel small molecule therapeutic.
Keywords: Pharmacological chaperone, protein homology modeling, ACT domain dimerization, protein multimerization, morpheein
Diminished activity of human phenylalanine hydroxylase (PAH), which catalyzes the iron, dioxygen, and BH4 dependent oxidation of phenylalanine (Phe) to tyrosine, is the most common inborn error of metabolism; the most severe cases result in phenylketonuria (PKU) [1]. Despite the success of dietary therapy in preventing severe PKU-associated mental deficits, patients continue to struggle with lifelong control of blood Phe levels. Poor control can compromise behavior, learning, and executive function [2–3]. Thus, alternative therapies are sought, including small molecule therapeutics [1, 4]. Pharmacological chaperones that stabilize PAH structure are gaining support as a long term therapeutic approach [5–12]. The cofactor BH4, which is an active site directed pharmacological chaperone, is in clinical use with some success [13]. Although biological and chemical chaperones are generally viewed as assisting in all aspects of protein folding, a focus on quaternary structure dynamics provides a different perspective that is applicable to PAH. Pharmacological chaperones can function allosterically (e.g. not at the enzyme active site) to assist a protein in achieving and/or maintaining its most active structure. PAH is known to be an allosteric enzyme where allostery involves major conformational changes and an equilibrium of multimers including dimers and tetramers. Although the structural basis for allosteric activation by Phe is poorly understood, PAH was recently identified as a putative morpheein, and the current work builds on this hypothesis [14–15]. Unlike previous views of PAH multimerization equilibria, the morpheein model contains two different PAH tetramers separated by a large kinetic barrier that requires multimer dissociation, conformational change in the dissociated state, and reassociation to an alternate tetramer. Herein we 1) detail the rationale behind developing a morpheein model for PAH allostery, 2) present protein structure models for components of a PAH quaternary structure equilibrium, and 3) provide experimental evidence on purified full length rat and human PAH consistent with the proposed equilibrium. The current work sets the stage for our long-term goal, which is to develop allosteric pharmacological chaperones to expand the PKU patient population responsive to small molecule therapy.
A quaternary structure equilibrium model for the dysfunction of PAH leading to PKU
A contemporary explanation for PAH dysfunction causing PKU identifies the disorder as one of PAH protein stability and/or folding [10, 12, 16–18]. For multimeric proteins, where the final step in the folding process is multimer assembly, perturbation of an equilibrium of functionally distinct alternative multimers provides both a mechanism for allostery as well as a mechanism for protein dysfunction [19–20]. Extensive literature describes mammalian PAH as an allosteric enzyme [21–22]. At low Phe levels the protein has basal activity which allows retention of sufficient Phe to support protein biosynthesis. At high Phe levels protein activation is accompanied by a large conformational change which dramatically changes various characteristics of the protein, including the fluorescence spectrum and the susceptibility to proteolytic digestion [23–25]. PAH allostery is associated with a quaternary structure equilibrium consisting of dimers and tetramers with some propensity to form larger aggregates [21, 23, 26–29]. Phe serves both as substrate and as allosteric activator; allosteric activation by Phe has been shown to draw an equilibrium of tetramers and dimers toward the tetramers [28, 30]. A structural basis for allosteric Phe binding causing stabilization of a specific, fully active conformation of the tetramer is proposed herein.
The new model for allosteric regulation of PAH, its relationship to PKU, and the promise of developing pharmacological chaperones that function allosterically follows our recently reviewed work with the prototype morpheein porphobilinogen synthase (PBGS) whose dysfunction causes the inborn error of metabolism known as ALAD porphyria [31–32]. Four important corollaries exist between the behavior of PBGS and the model proposed for PAH.
On the level of protein structure dynamics, the nature of the conformational change that dictates subunit assembly into either a high activity multimer or a low activity multimer is a rotation or hinge motion between two domains of each subunit. For human PAH, which is a three-domain protein detailed in Figure 1a, the proposed rotation is around the hinge at position 117, which lies between the regulatory and catalytic domains. The rationale for this proposal is based on an analysis of ACT domains in the Protein Data Bank (PDB) [33–36], described in more detail below. In support of the proposed PAH conformational change, hydrogen/deuterium exchange studies indicate that allosteric activation by Phe involves a dramatic reorientation of the regulatory and catalytic domains [37].
The structural commonality that imparts low activity on an assembly derives from protein-protein interactions that modulate molecular motions governing active site access. Figure 1b includes one orientation of the PAH domains, as seen in a rat PAH crystal structure (PDB id 1PHZ), where the N-terminal portion of the regulatory domain partially blocks active site access and is thus predicted to be the low activity conformation [38]. Reorientation of the entire regulatory domain would disallow this inhibitory interaction, placing the N-terminal portion out of reach of the enzyme active site.
The link between protein structure dynamics and disease is the effect of disease-associated mutations on the protein quaternary structure equilibrium. In the case of ALAD porphyria, which is a very rare disease, all eight disease-associated point mutations, which are dispersed throughout the protein structure, cause the quaternary structure equilibrium to be shifted toward the low activity assembly relative to the wild type protein [32]. With regard to PAH and PKU, there are hundreds of disease-associated point mutations throughout the protein structure [39], most of which are not at the enzyme active site, and some of which have already been demonstrated to affect an equilibrium between alternate multimers, though a model consisting of alternate tetramers has not previously been considered [16–17, 23–24, 27, 40–42]. We posit that disease-associated PAH mutations that shift a quaternary structure equilibrium will respond to pharmacological chaperones designed to act allosterically to stabilize the high activity multimer(s).
A strong precedent for the feasibility of discovering effective allosteric pharmacological chaperones is set by PBGS, where both in silico and in vitro screening methods have successfully identified small molecules that can perturb an equilibrium between low activity hexamers and high activity octamers and modulate enzyme activity [43–48]. The search for these allosteric regulators was based on oligomer-specific small-molecule binding sites. For PAH, the proposed structure for the high activity multimer has a protein-protein interface not present in the low activity multimer; portions of this interface are proposed as the oligomer-specific binding site for allosteric Phe. Other portions of this interface can be targeted for the development of pharmacologic chaperones.
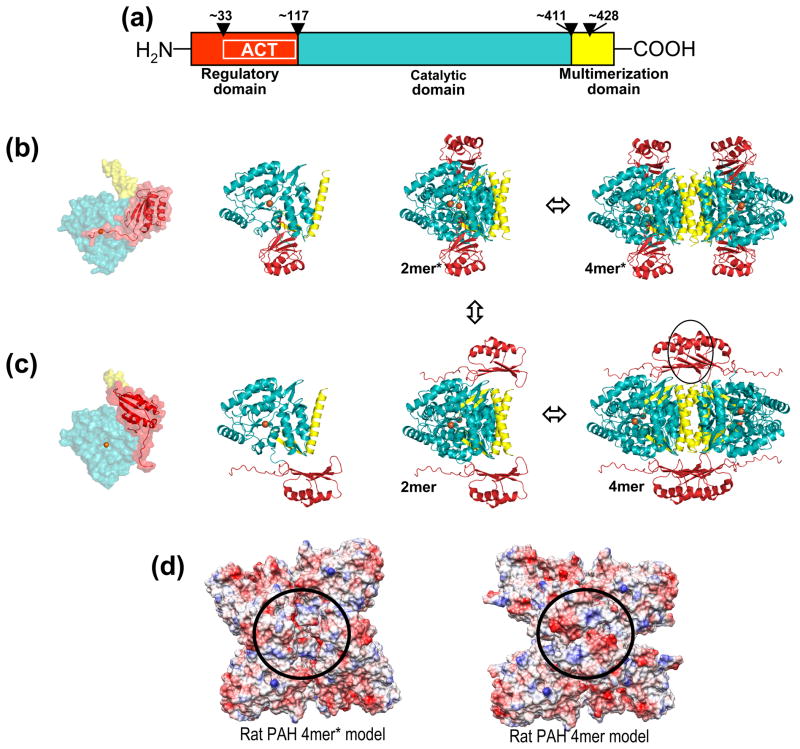
Figure 1. Mammalian PAH structure.
(a) The domain structure of mammalian PAH, numbered for the human protein. Wedges denote hinge regions. (b) On the left are two orientations of the human PAH monomer model, colored as in part a. The active site Fe is shown as an orange sphere. The transparent space filling representation illustrates the N-terminal region covering the active site. The catalytic and multimerization domain structures derive from PDB id 2PAH; the regulatory domain is a homology model based on the rat PAH structure PDBid 1PHZ, which contains both the regulatory and catalytic domains. The human PAH two-domain crystal structure is a tetramer; the corresponding tetramer for the three-domain model is shown on the right. This represents the low activity tetramer designated 4mer*. Also represented is the 2mer* ⇔ 4mer* equilibrium. (c) Following the organization of part b is the PAH model proposed to represent the high activity tetramer designated 4mer. The black circle represents the 4mer-specific subunit-subunit interaction. Also illustrated is the 2mer ⇔ 4mer equilibrium. An arrow connecting parts b and c completes the illustration of the 4mer* ⇔ 2mer* ⇔ 2mer ⇔ 4mer equilibrium. (d) Surface charge representations for similar models prepared for rat PAH. The circle shows the area of increased anionic character for 4mer relative to 4mer*.
The goals of this study are to prepare and support a model for the quaternary structure equilibrium that governs PAH activity and to develop methods that can evaluate the position of the PAH quaternary structure equilibrium in a variety of physiologically relevant environments. The developed methods will allow future investigation of the effect of PKU-associated variants on the position of the PAH quaternary structure equilibrium. The developed protein structure models will assist in pursuit of allosteric pharmacologic chaperones that promote PAH to achieve and maintain its active multimeric assembly.
MATERIALS & METHODS
Building the models
Both human PAH tetramer models are based on the human PAH structure 2PAH, which is a tetramer of a truncated form of PAH that contains only the catalytic and multimerization domains. A human regulatory domain homology model (residues 19–117) was prepared from the rat PAH crystal structure 1PHZ, which is of a truncated rat PAH containing only the regulatory and catalytic domains. Modeling was performed using our program MolIDE, which includes side chain rotamer optimization [49–50]. The low activity human PAH tetramer model was prepared using the program CHIMERA [51–52] to align the catalytic domains of the rat structure 1PHZ with the human structure 2PAH and the human regulatory domain model. Loop modeling (amino acids 113–121) was used to attach a regulatory domain to each of the human PAH subunits to yield our structure of 4mer*, using the program YASARA [53]. Following loop modeling, loop optimization included energy minimization by steepest descent methods. A final optimization of side chain rotamers was done in the context of the full multimer using SCWRL4 [54]. Preparation of the high activity human 4mer model started with the preparation of a dimer of the human PAH regulatory domain homology model, prepared by aligning two regulatory domains with the ACT domain dimer seen in crystal structure 1PSD. Following energy minimization and side chain optimization, the human PAH regulatory domain dimers were manually positioned to optimize the connection to the catalytic domains in the region of amino acid 118 of two catalytic domains with amino acid 117 of two halves of the regulatory domain homology model dimer. Loop modeling followed by side chain optimization and energy minimization completed the preparation of the human PAH 4mer model. Similar methods were used to prepare models of rat PAH 4mer* and 4mer.
Expressing and purifying the proteins
The expression construct for a the MBP fusion protein of human PAH was a kind gift of Dr. A. Martinez and protein purification followed standard procedures on an amylose column. Expression constructs for full length rat and human PAH were a kind gift of Prof. P. Fitzpatrick. All gifted plamids were amplified and the sequence was confirmed to be as published. Expression procedures included small variations on the procedures used for expression of mammalian PBGS [55]. Expression typically was carried out at reduced temperature (15 – 25 °C), Fe(II) was added at the same time as IPTG (at various concentrations), and in some cases glycerol was included as a chemical chaperone. None of these variations had a significant impact on protein expression levels. Purification of rat PAH followed the procedure of Shiman wherein crude cell extract was passed over a 75 ml Phenyl Sepharose-CL4B column in the presence of phenylalanine, followed by extensive washes with a variety of buffered solvents, and elution of the protein following removal of Phe from the elution buffer, which contains 15% glycerol, all carried out at 4 °C [56]. This affinity purification is based on a major conformational change that has long been known to accompany the process wherein Phe activates mammalian PAH [21]. Although this purification works well for rat PAH, human PAH, which expressed poorly, emerged from the column requiring significant further purification, which was carried out by ion exchange chromatography (IEC) on a 1 ml HiTrap Q column in 30 mM Tris-HCl, pH 7.4 using a KCl gradient (0–0.4 M) at room temperature.
Analytical chromatographic methods were carried out at room temperature on an AKTA protein purification system
Analytical SEC (100 μl sample size) was carried out on a 24 ml Superdex 200 column at 0.5 ml/min using 30 mM Tris pH 7.4, 150 mM KCl buffer with or without various concentrations of Phe. Analytical IEC was carried out on a 1 ml HiTrap Q column at a flow rate of 0.5 ml/min using 30 M Tris pH 7.4 with a KCl gradient from 20 mM to 0.4M with a variety of additives (see results).
Electrophoresis
Electrophoretic methods used the GE PhastSystem using 12.5% acrylamide gels and either native or SDS buffer strips. Western blotting on both native and denaturing gels used the WesternBreeze Chromogenic Western Blot Immunodetection Kit, WB7103 (Invitrogen). The primary antibody was MAB5278 (Millipore,10 ug/blot), which recognizes an epitope in the catalytic domain that is common to human tryptophan hydroxylase, tyrosine hydroxylase, and PAH.
Limited trypsin digestion
Rat PAH was subjected to limited proteolysis by trypsin (Roche sequencing grade). Trypsin stock solutions were prepared in 1 mM HCl, and the total amount of HCl added to each reaction was normalized. Digestion reactions were carried out at room temperature in phosphate buffered saline pH 7.4 with PAH held constant at 0.2 mg/mL. Ratios of trypsin:PAH were varied from 1:5 to 1:500, in the presence or absence of 1 mM Phe. At various times, aliquots of the digestion were removed and the digestion was terminated with 10 mM PMSF prior to resolution of the digestion products by SDS-PAGE.
PAH activity assays
PAH activity determination used a continuous fluorescence assay monitoring the production of tyrosine [57] adapted to single microcuvette format for use with a Photon Technologies International fluorescence spectrophotometer equipped with a xenon flash lamp. Fluorescence was monitored at λEx = 275 nm and λEm = 305 nm, with excitation and emission slit widths set to 2 nm. The standard assay (200 μL final volume) contained 20 mM Na-Hepes pH 7.3, 10 μM ferrous ammonium sulfate, 2 mM dithiothreitol, 1 mg/mL catalase (to yield 2000–5000 U/mL), 1 mM Phe, 75 μM BH4, and 1–5 μg PAH at 30 °C. For experiments monitoring “order-of-addition” effects, the assay components were allowed to equilibrate at 30 °C for 5 minutes (rat PAH) or 15 minutes (human PAH) prior to starting the reaction by addition of the final assay reaction component. Quantification of tyrosine was performed by constructing standard curves of tyrosine under assay solution conditions in the presence of the relevant concentrations of BH4, which can quench tyrosine fluorescence [57].
PAH tryptophan fluorescence emission spectra
Human and rat PAH were diluted to 0.2 mg/mL in 20 mM Na-Hepes pH 7.3, 2 mM DTT, 10 μM ferrous ammonium sulfate and 200 mM KCl in the presence or absence of 1 mM Phe. Emission spectra were recorded at 25 °C with λEx = 295 nm, and the excitation and emission slits set to 2 and 5 nm, respectively.
RESULTS
Protein structure models representing the PAH quaternary structure equilibrium
A protein structure model for the full length human PAH (Fig 1b, coordinates included in Supplementary Information) was prepared by combining 1) a homology model of the regulatory domain drawn from the crystal structure of a truncated rat PAH containing both the regulatory and catalytic domains (PDBid 1PHZ) with 2) the crystal structure of a truncated human PAH containing both the catalytic and multimerization domains (PDBid 2PAH). Similar straightforward combined models of full length tetrameric PAH have been presented previously [58]: they provide the standard representation of tetrameric PAH [22]. In these models the most N-terminal ordered region partially blocks access to the Fe-containing enzyme active site (Fig 1b). It is well documented that removal of the regulatory domain renders the protein constitutively active; this form is insensitive to Phe activation and has a specific activity comparable to the Phe-activated form of the full length protein [21]. Thus the tetramer shown in Fig 1b is assigned to the full length protein assembly with low or basal activity, which we designate 4mer*. The conformational change that differentiates the low activity from the high activity multimers is posited to disallow this inhibitory interaction between the regulatory and catalytic domains. A similar model for rat PAH 4mer*, where the multimerization domain is homology modeled, is included in Supplementary Information.
Formulating a structural basis for PAH allostery draws on the comprehensive review of Kaufman [21], which describes extensive biochemical, biophysical, and kinetic data on full-length human and rat PAH isolated from mammalian tissue. Although not previously interpreted in this framework, published data are largely consistent with a dissociative allosteric model as detailed for the morpheein model of allostery [14–15, 19, 59–60]. The proposed dissociative mechanism for allosteric regulation of mammalian PAH consists of an equilibrium of two structurally and functionally distinct PAH tetramers (4mer*, which is low activity, and 4mer, which is high activity) where the spatial relationship between the regulatory and catalytic domains changes dramatically. The required conformational change is proposed to be sterically hindered in the tetrameric states, and to occur at the level of a dimer, hypothetically through the pathway illustrated in Fig 1b & c. The structure of the activated tetramer is conceived to include a protein-protein interaction, which is not present in the low activity tetramer, and which provides a hypothetical binding site for an allosteric Phe. Allosteric Phe binding is supported by published Phe binding stoichiometry studies [61–62]. Based on the behavior of ACT domains, described in more detail below, allosteric Phe binding is proposed to occur at an interaction interface between the regulatory domains of two subunits (Fig 1c, coordinates included in Supplementary Information). This proposition is consistent with a recent biophysical characterization of the isolated rat PAH regulatory domain, which was unexpectedly discovered to be both dimeric and capable of binding Phe [63]. Consistent with our model, early work on the kinetic characteristics of dimeric and tetrameric mammalian PAH yielded the hypothesis that an allosteric Phe binding site is present in a tetramer and absent in the dimer [64]. According to the proposed equilibrium, 4mer* ⇔ 2mer* ⇔ 2mer ⇔ 4mer, allosteric Phe binding would stabilize the 4mer-specific interaction and draw the equilibrium toward 4mer. The active sites of 4mer, as modeled, have unfettered access to substrates (Fig 1c). Although the data presented herein do not address whether dissociation to the dimer is an essential component of the interconversion between the low activity tetramer (4mer*) and the high activity tetramer (4mer), we provide evidence for inclusion of a PAH dimer in the equilibrium as well as evidence for additional species in the equilibration of 4mer* with 4mer. A significant unknown in modeling the pathway between the tetramers is an uncertainty as to the conformation of the most N-terminal portion of the regulatory domain in the active 4mer (see below). Hydrogen/deuterium exchange studies show this region to be flexible both before and after Phe activation [37]. Indirectly supporting a role for dimers in this pathway, the same studies show significant flexibility in the C-terminal helical portion of the multimerization domain both in the presence and absence of the allosteric Phe.
A large portion of the PAH regulatory domain contains a sequence motif known as an ACT domain, named for three allosterically regulated enzymes containing this sequence (aspartate kinase, chorismate mutase, and TyrA (a.k.a. prephenate dehydrogenase)) [33, 35]. Modeling the structure of 4mer draws on the structure(s) of ACT domains in the PDB [34] as analyzed by ProtCID and PDBfam [65–66] as well as a recent review on the role of ACT domains in the regulation of amino acid and nucleoside metabolism [36]. Analysis of ACT domains in the PDB reveals that ACT domains commonly form dimers, as illustrated in Figure 2, where the ACT:ACT interface is often seen bound to a regulatory amino acid ligand. Publications on ACT domains often cite PAH as an outlier that does not include this interface [35–36]. However, applying this multimerization motif to the commonly accepted PAH tetramer structure, 4mer*, would require a ~90° rotation of the regulatory domain relative to the catalytic domain, not unlike the ~114° rotation seen in an ACT-like regulatory domain of 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase (DAH7PS) (see discussion) [67]. In building the high activity or allosterically activated PAH tetramer model, the initial step was to dimerize the human PAH regulatory domain homology model by forming an interface between the ACT domain segments. The ACT domain dimer seen in the crystal structure of phosphoglycerate dehydrogenase (PDBid 1PSD, Fig 2b) was used as a template for orienting the PAH ACT domains. The resultant regulatory domain dimer(s) were attached to the human PAH tetramer (PDB code 2PAH) at the hinge region that connects the regulatory and catalytic domains to yield the 4mer model illustrated in Figure 1c. One aspect of the 4mer model which has low confidence is the conformation of the N-terminal 33 amino acids and its orientation relative to the ACT domain; the model simply retains these structural elements from the rat PAH crystal structure. The coordinates for the human PAH 4mer model and a similarly prepared rat PAH 4mer model are both included in Supplementary Information.
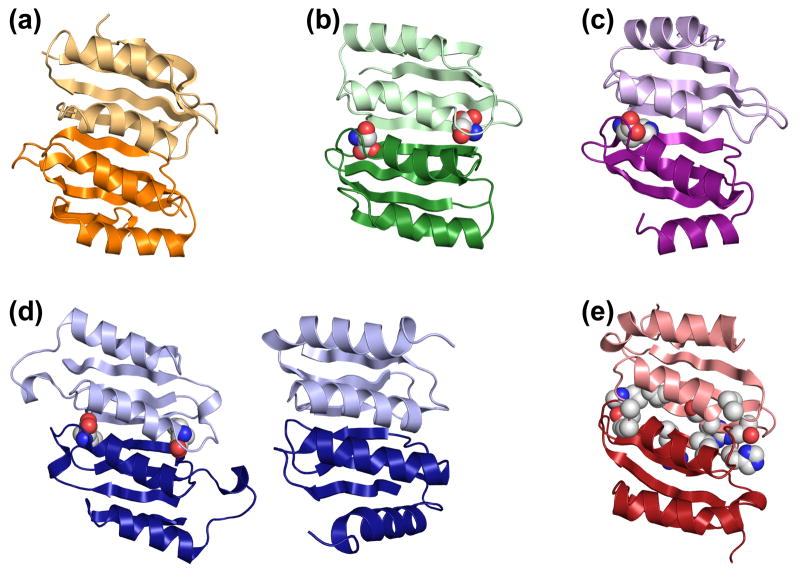
Figure 2. ACT domain dimer structures.
The ACT domain dimers are illustrated as ribbons with each subunit pair in light and dark shades. Using a space filling representation for the ligands bound at the ACT:ACT interface, examples are shown for (a) the regulatory subunit of acetolactate synthase, PDB id 2F1F, which has no ligand [76]; (b) phosphoglycerate dehydrogenase, PDB id 2PC9, which shows serine bound at the subunit-subunit interface [77]; (c) one of four ACT:ACT dimers present in protein BT0572 from Bacteroides thetaiotomicron, PDB id 2F06, where each ACT:ACT dimer in this structure contains one molecule of histidine at the subunit-subunit interface; and (d) asparate kinase, PDB id 2CDQ, which has two different (tandem) ACT:ACT dimers, only one of which contains lysine bound at the subunit-subunit interface [78]. (e) The ACT domains of human PAH in the active 4mer model. Shown in space filling representation are amino acid side chains that are substituted in disease-associated human PAH variants that have been shown to obliterate allosteric Phe binding [71].
Biochemical and biophysical data support the proposed model for PAH allostery
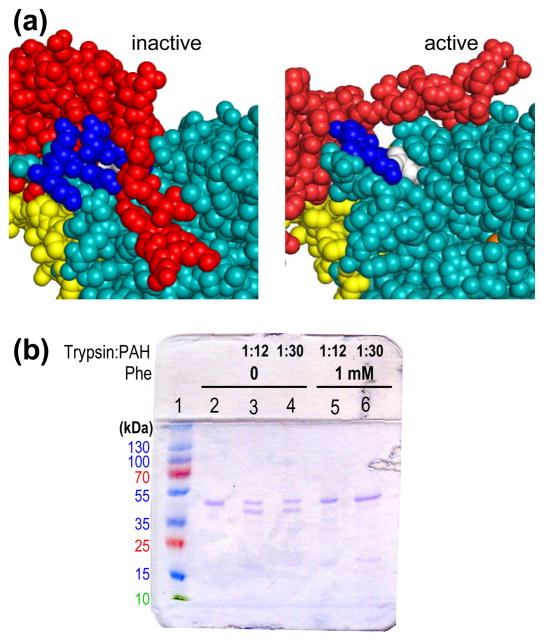
The models of 4mer* and 4mer are consistent with the well documented behaviors of PAH. For example, fluorescence ascribed to Trp120 and tryptic susceptibility in a highly basic region at the C-terminal region of the regulatory domain are two characteristics well documented to differentiate low activity PAH from Phe-activated PAH [23, 68–69]. Figure 3a illustrates that these regions of PAH are in different environments in 4mer relative to 4mer* consistent with published data. Using purified rat and human PAH, described below, we confirmed the 10 nm red shift in the PAH fluorescence spectra upon addition of sufficient Phe known to provide maximum activity stimulation (~1 mM Phe) [70]. We also show that the susceptibility of purified rat PAH to limited tryptic digestion is a function of whether or not Phe is present (Fig 3b). Under the conditions shown, 1 mM Phe provides nearly total protection from limited tryptic digestion, which has previously been shown to cleave within the basic region at the C-terminal end of the regulatory domain [69]. For rat PAH (Fig 3a) the surface accessibility of basic residues in this region decrease significantly when the conformation changes from the low activity to the high activity tetramers (e.g. for Arg111, Lys113, and Arg123, modeled surface accessibilities decrease to 69, 25, and 79 percent respectively). Hydrogen/deuterium exchange studies indicate that the entire interface between the regulatory and catalytic domains is more solvent exposed in the allosterically activated form, consistent with the 4mer (more exposed) and 4mer* (less exposed) models [37]. The proposed models also provide a structural explanation for some published data on disease-associated PAH variants. For example, ex post facto inspection of 4mer reveals that Gly46, Ala47, Thr63, His64, Ile65, and Arg68 correspond to the amino acid ligand binding sites seen in ACT domain dimers (compare Figs 2b, c, d, and e), and would thus be predicted to be in the vicinity of the allosteric Phe binding site on 4mer. Notably, and in strong support of our model for ACT domain dimerization forming the allosteric Phe binding site, in vitro characterization of PAH fusion proteins for the PKU-associated variants G46S, A47T, T63P/H64N, I65T, and R68S showed that these mutations eliminate Phe binding [71].
Figure 3. The proposed models are consistent with PAH behaviors.
(a) Shown are spacefill representations of a portion of rat PAH 4mer* (left) and 4mer (right) colored as in Figure 1, showing Trp120 in white and the highly basic C-terminal portion of the regulatory domain in blue. (b) SDS PAGE analysis of limited proteolysis of PS purified rat PAH (0.2 mg/mL) in the presence and absence of Phe for 1 hour at room temperature. Lanes: 1) molecular weight markers; 2) PAH incubated in the absence of Phe or trypsin; 3–6) PAH incubated in the absence or presence of Phe and trypsin as indicated.
In vitro characterization of purified PAH as a prerequisite to the characterization of disease-associated PAH variants
Although the proposed allosteric and structural models are consistent with published data on mammalian PAH, a unique aspect of the morpheein model is an equilibrium of two tetrameric assemblies separated by an unusual kinetic barrier which is proposed to include multimer dissociation, conformational change in the dissociated state, and assembly to an alternate multimer structure. In particular, the novel 4mer model suggests that the alternate tetramers can be separated using ion exchange chromatography (IEC); Fig 1d shows the enhanced anionic character of the 4mer surface relative to the 4mer* surface. IEC has been useful in distinguishing between the PBGS hexamer and octamer and in evaluating how disease associated variants shift the PBGS quaternary structure equilibrium [32]. We foresee IEC as a major tool in the future evaluation of disease-associated PAH variants. Using such classic biochemical techniques to establish an equilibrium of slowly exchanging protein quaternary structure isoforms can be straightforward if one presupposes the existence of such an equilibrium in the interpretation of experimental data. In addition to IEC, particularly useful and generally accessible experimental methods are native polyacrylamide gel electrophoresis (PAGE) and size exclusion chromatography (SEC). For an equilibrium of homo-multimers, native PAGE separates on the basis of multimerization stoichiometry because all multimers of a given protein have the same charge/mass ratio. SEC provides an independent estimate of multimerization stoichiometry (size). Both native PAGE and SEC can deviate from ideal behavior due to differences in shape, and such evidence is presented for PAH to support tetramers of significantly different shape (see below). Although these techniques are relatively low resolution, as described below, together they present compelling data in favor of a PAH quaternary structure equilibrium that consists of dimers and at least two architecturally different tetramers.
Experimental data has been obtained on mammalian PAH, heterologously expressed in Escherichia coli, from three different expression vectors. Because the human protein is reported to be unstable, most of the published work utilizes either a fusion protein or fragments lacking the regulatory domain. Below we describe studies of human PAH purified with a cleavable N-terminal maltose binding protein (MBP) as well as studies with full length human PAH expressed and purified without a tag. Fortunately, full length rat PAH is reported to be more tractable and expression, purification, and characterization of the tag-less rat protein informed our approach to working with tag-less full length human PAH. Our long-range goal of understanding how PKU-associated variants perturb a human PAH quaternary structure equilibrium demands establishing a baseline for the full length wild type human protein.
Human MBP-PAH
Initially human PAH was expressed as the MBP-PAH fusion and was found to largely follow published behavior [28], which is consistent with the proposed allosteric model. However, in the modeled fragile protein quaternary structure equilibrium, where the long-range plan is to evaluate disease-associated variants for their effect on said equilibrium, it is not reasonable to expect the MBP-PAH fusion to reflect a physiologically relevant equilibrium representative of full length PAH. For example, an N-terminal His-tag on DAH7PS has been shown to lock the position of its ACT-like regulatory domain and prevent its rotation [67]. Various conditions used to cleave MBP from the fusion protein resulted in either incomplete digestion or undesired nonspecific proteolysis yielding PAH dimers and tetramers that were heteromeric in size, surface charge, and charge/mass ratio, and thus unacceptable for the proposed methodology.
Shiman affinity purification of full length PAH
Full length (tag-less) rat and human PAH were expressed from published pET constructs with superior expression observed for the rat protein [72]. Following the Phenyl Sepharose affinity method of Shiman [56] the yield for rat PAH was ~12 mg at >90% purity (SDS PAGE in Fig 4a) from 9.1 g of E. coli cell paste with a specific activity of 7900 nmol min−1 mg−1 when addition of PAH is used to initiate activity. Expression of full length human PAH in E. coli used a variety of protocols, all of which provided relatively poor yield. Consequently 47.7 g of E. coli cell paste was used in the classic Shiman single column purification protocol. Unlike the behavior of rat PAH, human PAH activity eluted very slowly from the Phenyl Sepharose CL-4B (PS) column and the resulting PS pool measured 670 ml. This pool was concentrated to ~15 ml at ~ 1 mg/ml where SDS PAGE showed it to be of poor purity (Fig 4b) with a specific activity of only 1200 nmol min−1 mg−1. Nevertheless, an SDS Western blot yielded a single band for PAH (Fig 4b). A portion of the PS purified human PAH was further purified by IEC, described in more detail below, which yielded ~ 1mg of pure protein (Fig. 4b).
Figure 4. SDS PAGE analysis of rat and humanPAH.
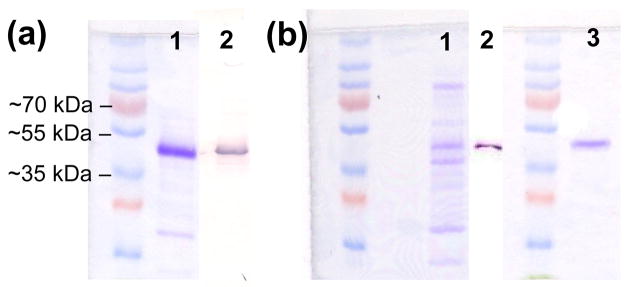
(a) PS purified rat PAH; lane 1 is Coomassie stained, lane 2 is a Western blot. (b) Human PAH; lane 1 is the concentrated PS pool (Coomassie stained); lane 2 is a Western blot of the concentrated PS pool; lane 3 is a Coomassie stained gel of IEC purified human PAH (pool A).
Order of addition effects on PAH catalyzed reaction rates
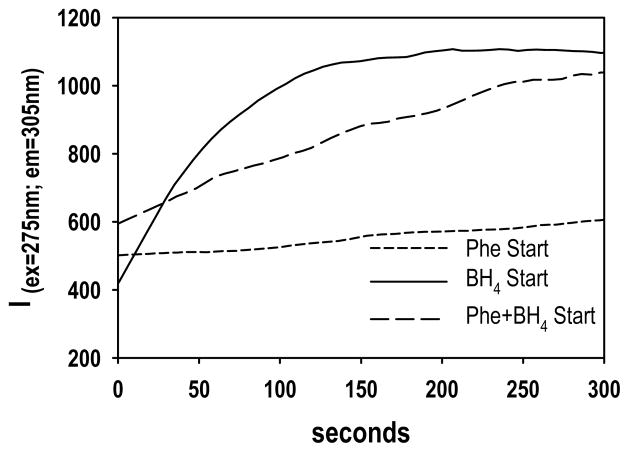
The PS purified rat PAH was used to illustrate dramatic differences in PAH catalyzed reaction rates during assays that vary only in the order-of-addition of reaction components; this kinetic behavior is one characteristic of morpheeins [60]. As illustrated in Figure 5, when the PAH assay is initiated by the addition of BH4, the initial rate is rapid and decays as O2 becomes limiting. When the assay is initiated by the addition of Phe, the initial rate is quite slow. An intermediate rate is seen when both Phe and BH4 are used to initiate the assay. One interpretation of these anomalies is that Phe stabilizes 4mer, yielding high initial activity, while BH4 (in the absence of Phe) stabilizes a form with low or basal activity, perhaps 4mer*. IEC purified human PAH showed nearly identical order of addition effects (not shown).
Figure 5. Order of addition effects on PAH activity.
The order of addition of reaction components has a profound effect on the initial activity of rat PAH under the conditions of [Phe] = 0.3 mM; [BH4] = 0.075 mM; PAH = 20 μg/ml. Protein was preincubated for 5 min prior to the addition of substrate(s). Activity axis directly reflects fluorescence due to formation of tyrosine.
Chromatographic behavior of mammalian PAH
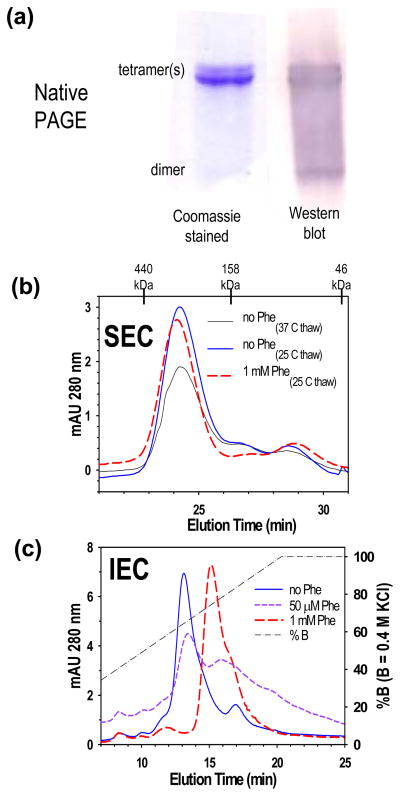
Considerable information on the behavior of PAH was derived from chromatographic and electrophoretic data. Native PAGE (Fig 6a) and SEC (Fig 6b) are consistent with the freshly purified rat PAH protein (at neutral pH with 15% glycerol) being predominantly tetrameric with a small amount of dimer; the dimeric component is more apparent in the Western blot relative to the Coomassie stained gel. Close inspection of the native PAGE results suggests two different rat PAH tetramers of different architecture. Close inspection of the SEC results suggests that rat PAH defrosted from −80 °C at 37 °C may contain more than one tetrameric component. This is not seen when the protein is defrosted at 25 °C, which is the temperature later used to defrost all other PAH samples. For comparison, a Coomassie stained SDS gel of freshly purified rat PAH, shown in Fig 4a, indicates a single band at the expected 53 kDa. To address whether or not IEC could resolve components of the quaternary structure equilibrium, purified rat PAH (~100 μg) was subjected to analytical IEC in a neutral pH buffer with a KCl gradient (Fig 6c) in the presence and absence of Phe. In the absence of Phe, the chromatogram showed predominantly one peak centered at 256 mM KCl. These same buffer conditions used for SEC analysis showed predominantly tetramer with a small amount of dimer (Fig 6b). Inclusion of 1 mM Phe in the IEC elution buffer, which is sufficient to fully activate rat PAH, dramatically altered the elution profile such that the main peak is centered at 294 mM KCl. The observed increased retention of rat PAH upon exposure to 1 mM Phe is consistent with the surface charge predicted for 4mer relative to 4mer* in rat PAH tetramer models (see Fig 1d). SEC (Figure 6b) indicates that 1 mM Phe does not dramatically alter the size of tetrameric PAH, although the precise elution of the PAH tetramer is slightly earlier in the presence of 1 mM Phe, consistent with the notion that 4mer* and 4mer have somewhat different shapes. The key demonstration that rat PAH exists in an equilibrium of multiple quaternary structure isoforms is its behavior on the analytical IEC column when 50 μM Phe is included in the elution buffers, but not the PAH sample; 50 μM Phe is sub-saturating for the allosteric activation of PAH. Under these conditions, rat PAH separates into both different tetramers, as well as additional forms (Fig 6c). At 50 μM Phe, rat PAH runs as an equilibrium of the form eluting at ~260 mM KCl (presumably 4mer*), the form eluting at ~300 mM KCl (presumably 4mer), as well as additional components that elute at higher KCl (~380 mM). The appearance of additional distinct chromatographically separable components is consistent with the participation of dimeric forms in the equilibration between alternate PAH tetramer structures.
Figure 6. Evidence for two different tetramers of rat PAH.
(a) PS-purified rat PAH is shown on native and native Western PAGE. (b) Analytical SEC (loading 100 μl at ~ 1 mg/ml) shows that there are not major changes in the tetrameric size of rat PAH in the presence or absence of Phe. (c) Analytical IEC (loading ~100 μg samples) under the same conditions as part b, reflects a significant change in the surface charge of rat PAH as a function of [Phe].
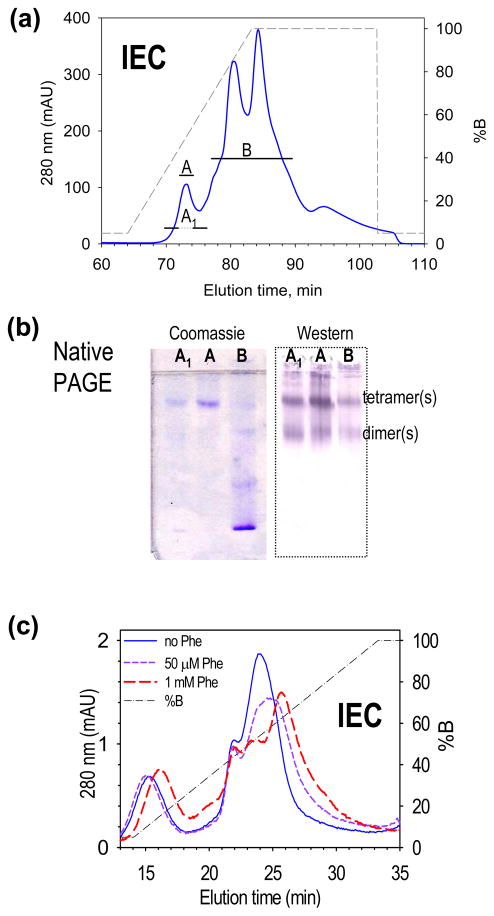
Using knowledge gained from the IEC behavior of rat PAH, the low-purity PS-purified human PAH was further purified by IEC, yielding two pools of activity (Fig 7a). The first peak, well resolved from the others, appeared pure by Coomassie stained SDS PAGE (Fig 4b) and of high specific activity. The fractions were brought to 15% glycerol and stored as the central fraction (Pool A, 6800 nmoles tyrosine min−1 mg−1), and the edge fractions (Pool A1, 6100 nmoles tyrosine min−1 mg−1). The region of disperse low activity (Pool B) coeluted with other proteins and was of lower specific activity (700 nmoles tyrosine min−1 mg−1). Coomassie stained native PAGE and native Western blots of these Pools are illustrated in Fig 7b, which shows that although Pool A is pure PAH (see Fig 4b), it contains tetramer, dimer, and some higher order PAH multimers that barely enter the gel (see Discussion).
Figure 7. IEC purification and analysis of human PAH.
(a) Further purification of PS purified human PAH by IEC yields a high-purity high-activity pool (A), and a disperse, low-purity low-activity pool (B). (b) Native PAGE and native Western indicate that each pool contains human PAH as both tetramer and dimer. (c) IEC behavior of 100 μl aliquots of pool A in the presence and absence of Phe at both 1 mM and 50 μM (compare to Fig 6c).
In order to investigate whether the IEC-resolved peaks of human PAH are in equilibrium, both Pools A1 and B were dialyzed against the PS elution buffer, and individually re-chromatographed by IEC. In case of Pool A1, rechromatography resulted in several peaks, notably one at 85 minutes. In the case of Pool B, rechromatography resulted in the reemergence of the original Pool B peaks plus a small peak at 74 min. The combination of these analyses supports the conclusion that the form of human PAH that elutes at 74 min is in equilibrium with a form of human PAH that elutes at 85 min. SDS PAGE (not shown) indicates that the protein subunits do not change molecular mass during this equilibration.
IEC purified human PAH (Pool A) was used to evaluate the effect of Phe on IEC mobility (Fig 7c). In this experiment 100 μl aliquots of protein were diluted to 1 ml in the presence of Phe at 0, 50 μM, and 1 mM, followed by a 1 hour incubation prior to IEC; the results showed the main peak of human PAH migrating at 214 mM KCl in the absence of Phe and at 247 mM KCl in the presence of 1 mM Phe. At 50 μM Phe a broad peak of intermediate mobility is seen. Without the incubation, the chromatographic changes were attenuated. Although interpretation of human PAH IEC chromatograms is not as clear-cut as a similar study done with rat PAH (Fig 6c), the data indicate that the Phe induced conformational change causes the protein to be further retained on an anion exchange resin.
Phe stabilization of PAH tetramer
To demonstrate that Phe stabilizes a PAH tetramer relative to the dimer(s), the predominantly tetrameric rat PAH (~ 1 mg/ml, Fig 6a), was incubated at ~ 0.2 mg/ml in the presence and absence of ligands. Under these conditions, time points (not shown) indicate that the protein slowly equilibrates between tetramer and dimer, yielding about a 50/50 mixture after 24 h at 25 °C (Fig 8). Phe at 1 mM provided complete stabilization of a tetramer, presumably 4mer; Phe at 50 μM did not appreciably stabilize the tetramer; BH4 at 75 μM appears to promote formation of a dimer; and this effect is modulated in the presence of both 75 μM BH4 and 50 μM Phe.
Figure 8. Phe stabilization of tetrameric rat PAH.
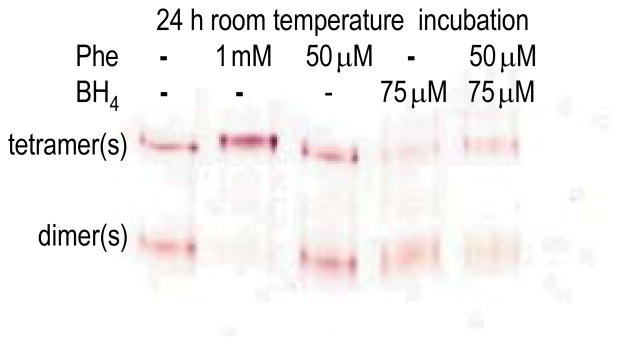
Native Western showing the effects of BH4 and Phe on the spontaneous tetramer(s)-dimer(s) equilibration of 0.2 mg/ml rat PAH.
DISCUSSION
We demonstrate for the first time that mammalian PAH exists in an equilibrium consisting of tetramers of two distinct architectures and that one or more dimeric forms of PAH are part of the equilibrium. The published behavior of mammalian PAH is consistent with a dissociative allosteric mechanism [21–22], which is approximated by a morpheein model of PAH allostery consisting of dimers and tetramers in the equilibrium 4mer* ⇔ 2mer* ⇔ 2mer ⇔ 4mer; 4mer is the most active assembly and has a distinct allosteric binding site for Phe that is not present in the other components of the equilibrium. 4mer stabilization is proposed as the mechanism wherein Phe acts as an allosteric activator of mammalian PAH. In 4mer the N-terminal portion of the regulatory domain cannot sample the active-site-blocking conformation illustrated for 4mer* and seen in the rat PAH two-domain crystal structure. Although the current work does not disallow a direct equilibration between the tetramers, e.g. 4mer* ⇔ 4mer, which would complete a thermodynamic box, we propose this interconversion to be sterically forbidden. We are developing the necessary tools to address this question directly as we have done for PBGS [59, 73]. A key to this demonstration is the isolation of a heterologously expressed human PAH variant whose quaternary structure equilibrium is dramatically shifted, as we found in the F12L variant of PBGS. Such variants are expected to emerge from the planned characterization of select disease associated PAH proteins.
In formulating further hypotheses about the PAH quaternary structure equilibrium, we look to the behavior of human PBGS, where a morpheein model for allostery is 6mer* ⇔ 2mer* ⇔ 2mer ⇔ 8mer, and the high activity 8mer is a fragile assembly [31]. The human PBGS protein assembly defaults to the low activity 6mer* in response to a variety of specific point mutations, to basic pH, and to the absence of active site ligands [31]. In fact the basic arm of the bell-shaped pH activity profile is attributed to a shift in the equilibrium from predominantly high activity 8mer to predominantly low activity 6mer* [74]. Specifically, all of the point mutations that encode variants associated with the inborn error of metabolism ALAD porphyria result in protein with an increased propensity to sample the hexameric assembly [32]. A variety of small molecule ligands have been described as acting at the active site as well as allosterically to modulate the human PBGS quaternary structure equilibrium and thus modulate PBGS activity [43–44, 47–48]. A plethora of literature on mammalian PAH suggests comparable behavior wherein a variety of factors such as pH and ligand binding are reported to affect enzyme activity and protein stability. Here we consider that the reported inability of some disease associated PAH variants to achieve an active conformation may reflect an altered quaternary structure equilibrium rather than a failure to properly fold the individual subunits. Accumulation of the low activity components of the 4mer* ⇔ 2mer* ⇔ 2mer ⇔ 4mer equilibrium is not an accumulation of improperly folded PAH; it is an accumulation of protein in a physiologically meaningful state with basal activity. Although we cannot predict the percentage of PKU-associated mutations that shift the PAH quaternary structure equilibrium, based on our experience with small molecule modulation of the PBGS quaternary structure equilibrium, we are confident that small molecules will be found that effectively shift the equilibrium toward the active 4mer.
PAH aggregation
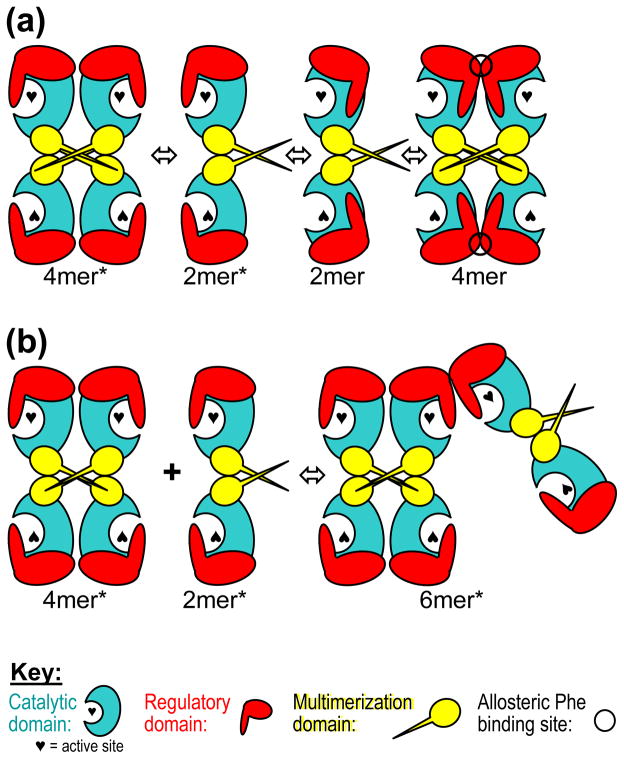
It is well documented that PAH has a tendency to “aggregate”, or to form higher order multimers, and we see evidence for this in our native Western blots (e.g. Fig 7b). The formation of 4mer from two 2mers, stabilized by ACT domain dimerization, forms a closed system without the ability to further multimerize. This is illustrated schematically in Figure 9a. However, one can model the observed higher order multimerization through the interaction between ACT domains of the 4mer* and 2mer* components of the PAH quaternary structure equilibrium. The association of ACT domains between 4mer* and/or 2mer* components always leaves an exposed ACT domain interaction surface that can further multimerize (Fig 9b). It is unknown if these higher order multimers exist in vivo or if they have any meaningful place in PAH allostery. Other multimeric allosteric metabolic enzymes such as inosine monophosphate dehydrogenase and CTP synthase have been shown to form higher order multimers that appear to function meaningfully in aspects of cell cycle control [75].
Figure 9. PAH multimerization schemes.
(a) A schematic of the proposed morpheein model for the allosteric regulation of PAH; formation of 4mer is a closed assembly unable to further multimerize. (b) Formation of higher order aggregates can occur through the dimerization of PAH regulatory domains when the individual subunits are in the 2mer* or 4mer* assembly. Formation of a 6mer* is illustrated. These are not closed assemblies and can further multimerize to larger assemblies through interaction of the multimerization domains and regulatory domains to form 8mer*, 10mer*, 12mer*, etc.
Thermatoga maritima DAH7PS, a fascinating correlation
DAH7PS (E.C. 2.5.1.54) catalyzes the first step in de novo aromatic amino acid biosynthesis via the shikimate pathway. DAH7PS, like PAH and PBGS, has evolved a phylogenetically diverse repertoire of mechanisms for allosteric regulation; for DAH7PS the regulation appears to be a classic feedback inhibition by tyrosine or phenylalanine. T. maritima DAH7PS is a homotetramer in which the structural basis for feedback inhibition was recently revealed to involve a large rotation (~114°) of an ACT-like regulatory domain relative to a catalytic domain [67]. Similar to the proposed allosteric regulation of PAH, the DAH7PS domain reorientation serves to gate access to the enzyme active site; this is not unlike the PBGS case wherein allosteric regulation modulates the opening/closing of an active site lid. In all three cases, the fundamental difference between the low activity state and the high activity state lies in the modulation of active site access, not in any repositioning of active site residues. Strikingly similar to the proposed PAH regulatory model, allosteric regulation of T. maritima DAH7PS involves dimerization of the ACT-like domain and tyrosine is seen bound at the domain interface (Fig S1). Strikingly dissimilar is the dimerization mode (Fig S1). However, unlike PBGS and PAH, there is no evidence to date that allosteric regulation T. maritima DAH7PS involves tetramer dissociation.
Supplementary Material
HIGHLIGHTS.
A new morpheein model for PAH allostery is proposed: 4mer* ⇔ 2mer* ⇔ 2mer ⇔ 4mer.
Protein structure models are presented for low activity 4mer* and Phe activated 4mer.
Mammalian PAH is shown to consist of an equilibrium of architecturally distinct multimers.
Spectroscopic, chromatographic, and kinetic data support the proposed models.
This work holds promise for developing allosteric pharmacologic chaperones to treat PKU.
Acknowledgments
The authors gratefully acknowledge support from the Developmental Therapeutics program at the Fox Chase Cancer Center, from the National Institutes of Health, National Cancer Institute Comprehensive Cancer Center grant P30CA006927, and the National Institute of General Medical Sciences R01GM84453 (RLD). We acknowledge contributions from the FCCC Molecular Modeling Facility, the Glasswashing Facility, and the DNA Sequencing Facility.
ABBREVIATIONS
- DAH7PS
3-deoxy-D-arabino-heptulosonate 7-phosphate synthase
- IEC
ion exchange chromatography
- MBP
maltose binding protein
- PS
phenyl Sepharose
- Phe
phenylalanine
- PAH
phenylalanine hydroxylase
- PKU
phenylketonuria
- PAGE
polyacrylamide gel electrophoresis
- PBGS
porphobilinogen synthase
- PDB
protein data bank
- SEC
size exclusion chromatography
- SDS
sodium dodecyl sulfate
- BH4
tetrahydrobiopterin
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Blau N, van Spronsen FJ, Levy HL. Lancet. 2010;376:1417–1427. doi: 10.1016/S0140-6736(10)60961-0. [DOI] [PubMed] [Google Scholar]
- 2.Antshel KM. Mol Genet Metab. 2010;99(Suppl 1):S52–58. doi: 10.1016/j.ymgme.2009.09.013. [DOI] [PubMed] [Google Scholar]
- 3.Gentile JK, Ten Hoedt AE, Bosch AM. Mol Genet Metab. 2010;99(Suppl 1):S64–67. doi: 10.1016/j.ymgme.2009.10.183. [DOI] [PubMed] [Google Scholar]
- 4.van Spronsen FJ, Enns GM. Mol Genet Metab. 2010;99(Suppl 1):S90–95. doi: 10.1016/j.ymgme.2009.10.008. [DOI] [PubMed] [Google Scholar]
- 5.Torreblanca R, Lira-Navarrete E, Sancho J, Hurtado-Guerrero R. Chembiochem. 2012 doi: 10.1002/cbic.201200188. [DOI] [PubMed] [Google Scholar]
- 6.Staudigl M, Gersting SW, Danecka MK, Messing DD, Woidy M, Pinkas D, Kemter KF, Blau N, Muntau AC. Hum Mol Genet. 2011;20:2628–2641. doi: 10.1093/hmg/ddr165. [DOI] [PubMed] [Google Scholar]
- 7.Leandro J, Saraste J, Leandro P, Flatmark T. Mol Genet Metab. 2011;104(Suppl):S40–44. doi: 10.1016/j.ymgme.2011.07.024. [DOI] [PubMed] [Google Scholar]
- 8.Muntau AC, Gersting SW. J Inherit Metab Dis. 2010;33:649–658. doi: 10.1007/s10545-010-9185-4. [DOI] [PubMed] [Google Scholar]
- 9.Gersting SW, Lagler FB, Eichinger A, Kemter KF, Danecka MK, Messing DD, Staudigl M, Domdey KA, Zsifkovits C, Fingerhut R, Glossmann H, Roscher AA, Muntau AC. Hum Mol Genet. 2010;19:2039–2049. doi: 10.1093/hmg/ddq085. [DOI] [PubMed] [Google Scholar]
- 10.Pey AL, Ying M, Cremades N, Velazquez-Campoy A, Scherer T, Thony B, Sancho J, Martinez A. J Clin Invest. 2008;118:2858–2867. doi: 10.1172/JCI34355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nascimento C, Leandro J, Tavares de Almeida I, Leandro P. Protein J. 2008;27:392–400. doi: 10.1007/s10930-008-9149-9. [DOI] [PubMed] [Google Scholar]
- 12.Martinez A, Calvo AC, Teigen K, Pey AL. Prog Mol Biol Transl Sci. 2008;83:89–134. doi: 10.1016/S0079-6603(08)00603-X. [DOI] [PubMed] [Google Scholar]
- 13.Blau N, Hennermann JB, Langenbeck U, Lichter-Konecki U. Mol Genet Metab. 2011;104(Suppl):S2–9. doi: 10.1016/j.ymgme.2011.08.017. [DOI] [PubMed] [Google Scholar]
- 14.Selwood T, Jaffe EK. Archives of Biochemistry and Biophysics. 2012;519:131–143. doi: 10.1016/j.abb.2011.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jaffe EK. Open Conf Proc J. 2010;1:1–6. doi: 10.2174/2210289201001010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leandro J, Simonsen N, Saraste J, Leandro P, Flatmark T. Biochim Biophys Acta. 2011;1812:106–120. doi: 10.1016/j.bbadis.2010.09.015. [DOI] [PubMed] [Google Scholar]
- 17.Gersting SW, Kemter KF, Staudigl M, Messing DD, Danecka MK, Lagler FB, Sommerhoff CP, Roscher AA, Muntau AC. Am J Hum Genet. 2008;83:5–17. doi: 10.1016/j.ajhg.2008.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gamez A, Perez B, Ugarte M, Desviat LR. J Biol Chem. 2000;275:29737–29742. doi: 10.1074/jbc.M003231200. [DOI] [PubMed] [Google Scholar]
- 19.Jaffe EK. Trends in Biochemical Sciences. 2005;30:490–497. doi: 10.1016/j.tibs.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 20.Gronenborn AM. Curr Opin Struct Biol. 2009;19:39–49. doi: 10.1016/j.sbi.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaufman S. Adv Enzymol Relat Areas Mol Biol. 1993;67:77–264. doi: 10.1002/9780470123133.ch2. [DOI] [PubMed] [Google Scholar]
- 22.Fitzpatrick PF. Arch Biochem Biophys. 2012;519:194–201. doi: 10.1016/j.abb.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bjorgo E, de Carvalho RM, Flatmark T. Eur J Biochem. 2001;268:997–1005. doi: 10.1046/j.1432-1327.2001.01958.x. [DOI] [PubMed] [Google Scholar]
- 24.Bjorgo E, Knappskog PM, Martinez A, Stevens RC, Flatmark T. Eur J Biochem. 1998;257:1–10. doi: 10.1046/j.1432-1327.1998.2570001.x. [DOI] [PubMed] [Google Scholar]
- 25.Flatmark T, Stevens RC. Chem Rev. 1999;99:2137–2160. doi: 10.1021/cr980450y. [DOI] [PubMed] [Google Scholar]
- 26.Kowlessur D, Citron BA, Kaufman S. Arch Biochem Biophys. 1996;333:85–95. doi: 10.1006/abbi.1996.0367. [DOI] [PubMed] [Google Scholar]
- 27.Leandro J, Leandro P, Flatmark T. Biochim Biophys Acta. 2011;1812:602–612. doi: 10.1016/j.bbadis.2011.02.001. [DOI] [PubMed] [Google Scholar]
- 28.Martinez A, Knappskog PM, Olafsdottir S, Doskeland AP, Eiken HG, Svebak RM, Bozzini M, Apold J, Flatmark T. Biochem J. 1995;306(Pt 2):589–597. doi: 10.1042/bj3060589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaufman S. Adv Enzymol Relat Areas Mol Biol. 1971;35:245–319. doi: 10.1002/9780470122808.ch6. [DOI] [PubMed] [Google Scholar]
- 30.Chestkov VV, Kovalev LI, Shishkin SS, Annenkov GA. Vopr Med Khim. 1985;31:60–65. [PubMed] [Google Scholar]
- 31.Jaffe EK, Lawrence SH. Archives of Biochemistry and Biophysics. 2012;519:144–153. doi: 10.1016/j.abb.2011.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jaffe EK, Stith L. American Journal of Human Genetics. 2007;80:329–337. doi: 10.1086/511444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aravind L, Koonin EV. J Mol Biol. 1999;287:1023–1040. doi: 10.1006/jmbi.1999.2653. [DOI] [PubMed] [Google Scholar]
- 34.Berman H, Henrick K, Nakamura H, Markley JL. Nucleic Acids Res. 2007;35:D301–D303. doi: 10.1093/nar/gkl971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chipman DM, Shaanan B. Curr Opin Struct Biol. 2001;11:694–700. doi: 10.1016/s0959-440x(01)00272-x. [DOI] [PubMed] [Google Scholar]
- 36.Liberles JS, Thorolfsson M, Martinez A. Amino Acids. 2005;28:1–12. doi: 10.1007/s00726-004-0152-y. [DOI] [PubMed] [Google Scholar]
- 37.Li J, Dangott LJ, Fitzpatrick PF. Biochemistry. 2010;49:3327–3335. doi: 10.1021/bi1001294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kobe B, Jennings IG, House CM, Michell BJ, Goodwill KE, Santarsiero BD, Stevens RC, Cotton RG, Kemp BE. Nat Struct Biol. 1999;6:442–448. doi: 10.1038/8247. [DOI] [PubMed] [Google Scholar]
- 39.Scriver CR, Hurtubise M, Konecki D, Phommarinh M, Prevost L, Erlandsen H, Stevens R, Waters PJ, Ryan S, McDonald D, Sarkissian C. Hum Mutat. 2003;21:333–344. doi: 10.1002/humu.10200. [DOI] [PubMed] [Google Scholar]
- 40.Thorolfsson M, Teigen K, Martinez A. Biochemistry. 2003;42:3419–3428. doi: 10.1021/bi034021s. [DOI] [PubMed] [Google Scholar]
- 41.Song F, Qu Y, Okano Y, Ye Z, Zhang Y, Jin Y, Wang H. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2008;25:1–5. [PubMed] [Google Scholar]
- 42.Cerreto M, Cavaliere P, Carluccio C, Amato F, Zagari A, Daniele A, Salvatore F. Biochim Biophys Acta. 2011;1812:1435–1445. doi: 10.1016/j.bbadis.2011.07.012. [DOI] [PubMed] [Google Scholar]
- 43.Lawrence SH, Selwood T, Jaffe EK. Current Chemical Biology. 2013;7(2) doi: 10.2174/2212796811307020011. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lawrence SH, Selwood T, Jaffe EK. ChemMedChem. 2011;6:1067–1073. doi: 10.1002/cmdc.201100009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reitz AB, Ramirez UD, Stith L, Du Y, Smith GR, Jaffe EK. ARKIVOC. 2010;2010:175–188. [PMC free article] [PubMed] [Google Scholar]
- 46.Ramirez UD, Myachina F, Stith L, Jaffe EK. Adv Exp Med Biol. 2010;680:481–488. doi: 10.1007/978-1-4419-5913-3_54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lawrence SH, Ramirez UD, Selwood T, Stith L, Jaffe EK. J Biol Chem. 2009;284:35807–35817. doi: 10.1074/jbc.M109.026294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lawrence SH, Ramirez UD, Tang L, Fazliyez F, Kundrat L, Markham GD, Jaffe EK. Chem Biol. 2008;15:586–596. doi: 10.1016/j.chembiol.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Canutescu AA, Dunbrack RL., Jr Bioinformatics. 2005;21:2914–2916. doi: 10.1093/bioinformatics/bti438. [DOI] [PubMed] [Google Scholar]
- 50.Wang Q, Canutescu AA, Dunbrack RL., Jr Nat Protoc. 2008;3:1832–1847. doi: 10.1038/nprot.2008.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 52.Yang Z, Lasker K, Schneidman-Duhovny D, Webb B, Huang CC, Pettersen EF, Goddard TD, Meng EC, Sali A, Ferrin TE. J Struct Biol. 2012;179:269–278. doi: 10.1016/j.jsb.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Krieger E, Joo K, Lee J, Raman S, Thompson J, Tyka M, Baker D, Karplus K. Proteins. 2009;77(Suppl 9):114–122. doi: 10.1002/prot.22570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krivov GG, Shapovalov MV, Dunbrack RL., Jr Proteins. 2009;77:778–795. doi: 10.1002/prot.22488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tang L, Breinig S, Stith L, Mischel A, Tannir J, Kokona B, Fairman R, Jaffe EK. Journal of Biological Chemistry. 2006;281:6682–6690. doi: 10.1074/jbc.M511134200. [DOI] [PubMed] [Google Scholar]
- 56.Shiman R, Gray DW, Pater A. J Biol Chem. 1979;254:11300–11306. [PubMed] [Google Scholar]
- 57.Gersting SW, Staudigl M, Truger MS, Messing DD, Danecka MK, Sommerhoff CP, Kemter KF, Muntau AC. J Biol Chem. 2010;285:30686–30697. doi: 10.1074/jbc.M110.124016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zurfluh MR, Zschocke J, Lindner M, Feillet F, Chery C, Burlina A, Stevens RC, Thony B, Blau N. Hum Mutat. 2008;29:167–175. doi: 10.1002/humu.20637. [DOI] [PubMed] [Google Scholar]
- 59.Jaffe EK, Lawrence SH. Methods Mol Biol. 2012;796:217–231. doi: 10.1007/978-1-61779-334-9_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lawrence SH, Jaffe EK. Biochem Mol Biol Educ. 2008;36:274–283. doi: 10.1002/bmb.20211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Parniak MA, Kaufman S. J Biol Chem. 1981;256:6876–6882. [PubMed] [Google Scholar]
- 62.Shiman R, Xia T, Hill MA, Gray DW. J Biol Chem. 1994;269:24647–24656. [PubMed] [Google Scholar]
- 63.Li J, Ilangovan U, Daubner SC, Hinck AP, Fitzpatrick PF. Arch Biochem Biophys. 2011;505:250–255. doi: 10.1016/j.abb.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Parniak MA, Kaufman S. Biochemistry. 1985;24:3379–3379. [Google Scholar]
- 65.Xu Q, Dunbrack RL., Jr Nucleic Acids Res. 2011;39:D761–770. doi: 10.1093/nar/gkq1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xu Q, Dunbrack RL., Jr Bioinformatics. 2012 doi: 10.1093/bioinformatics/bts533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cross PJ, Dobson RC, Patchett ML, Parker EJ. J Biol Chem. 2011;286:10216–10224. doi: 10.1074/jbc.M110.209924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Knappskog PM, Haavik J. Biochemistry. 1995;34:11790–11799. doi: 10.1021/bi00037a017. [DOI] [PubMed] [Google Scholar]
- 69.Stokka AJ, Carvalho RN, Barroso JF, Flatmark T. J Biol Chem. 2004;279:26571–26580. doi: 10.1074/jbc.M400879200. [DOI] [PubMed] [Google Scholar]
- 70.Shiman R, Jones SH, Gray DW. J Biol Chem. 1990;265:11633–11642. [PubMed] [Google Scholar]
- 71.Gjetting T, Petersen M, Guldberg P, Guttler F. Mol Genet Metab. 2001;72:132–143. doi: 10.1006/mgme.2000.3118. [DOI] [PubMed] [Google Scholar]
- 72.Daubner SC, Hillas PJ, Fitzpatrick PF. Arch Biochem Biophys. 1997;348:295–302. doi: 10.1006/abbi.1997.0435. [DOI] [PubMed] [Google Scholar]
- 73.Tang L, Stith L, Jaffe EK. Journal of Biological Chemistry. 2005;280:15786–15793. doi: 10.1074/jbc.M500218200. [DOI] [PubMed] [Google Scholar]
- 74.Selwood T, Tang L, Lawrence SH, Anokhina Y, Jaffe EK. Biochemistry. 2008;47:3245–3257. doi: 10.1021/bi702113z. [DOI] [PubMed] [Google Scholar]
- 75.Carcamo WC, Satoh M, Kasahara H, Terada N, Hamazaki T, Chan JY, Yao B, Tamayo S, Covini G, von Muhlen CA, Chan EK. PLoS One. 2011;6:e29690. doi: 10.1371/journal.pone.0029690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kaplun A, Vyazmensky M, Zherdev Y, Belenky I, Slutzker A, Mendel S, Barak Z, Chipman DM, Shaanan B. J Mol Biol. 2006;357:951–963. doi: 10.1016/j.jmb.2005.12.077. [DOI] [PubMed] [Google Scholar]
- 77.Dey S, Grant GA, Sacchettini JC. J Biol Chem. 2005;280:14892–14899. doi: 10.1074/jbc.M414489200. [DOI] [PubMed] [Google Scholar]
- 78.Mas-Droux C, Curien G, Robert-Genthon M, Laurencin M, Ferrer JL, Dumas R. Plant Cell. 2006;18:1681–1692. doi: 10.1105/tpc.105.040451. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.