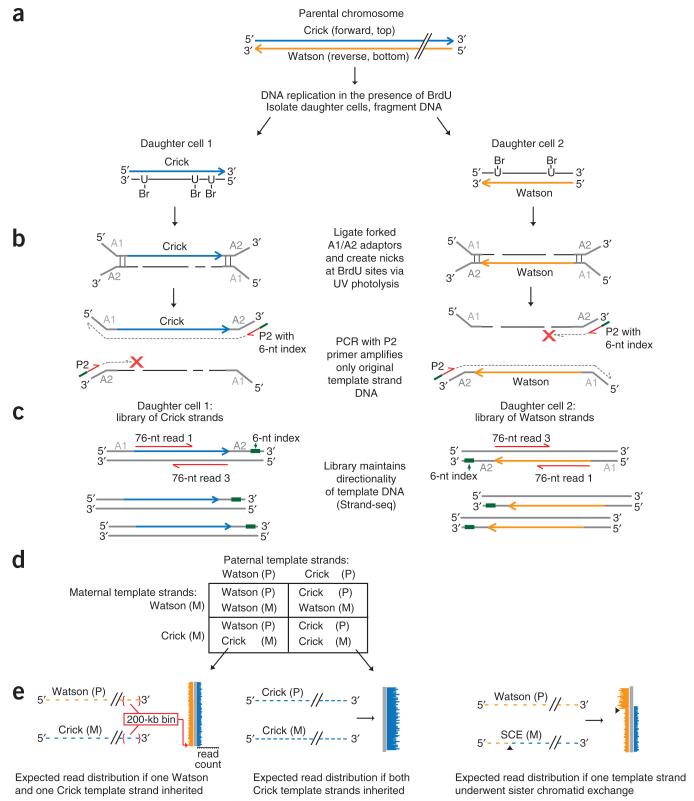
Figure 1.
Principle of single-cell DNA template strand sequencing. (a) Top: a single parental chromosome before DNA synthesis is shown with the Crick (blue) and Watson (orange) strands. Bottom: following DNA replication in the presence of BrdU, each sister chromatid with one original template strand and one complementary strand containing BrdU will segregate into one of the daughter cells. (b) DNA is fragmented and ligated to universal forked adaptors; UV photolysis creates nicks at BrdU sites, preventing PCR amplification of newly formed strands but allowing amplification of the original intact template strand. (c) The resulting libraries are directional, containing the template strand in its original genomic orientation in all amplified fragments. Multiple single-cell libraries containing unique 6-nucleotide (nt) index sequences (green lines in b) are pooled and sequenced on an Illumina platform. Two 76-nt reads from both directions (red lines) will read both the original template strand (always read 1) and the complementary strand (always read 3). (d) Possible combinations of maternal (M) and paternal (P) template strands inherited by daughter cells. (e) Expected read distribution from diploid Strand-seq libraries from inbred mouse cells. Watson and Crick reads (orange and blue lines) are binned and mapped to either side of a chromosome ideogram (gray). SCEs are expected to show a switch from both Watson and Crick reads to either Watson or Crick alone (right; arrowheads indicate SCE interval).