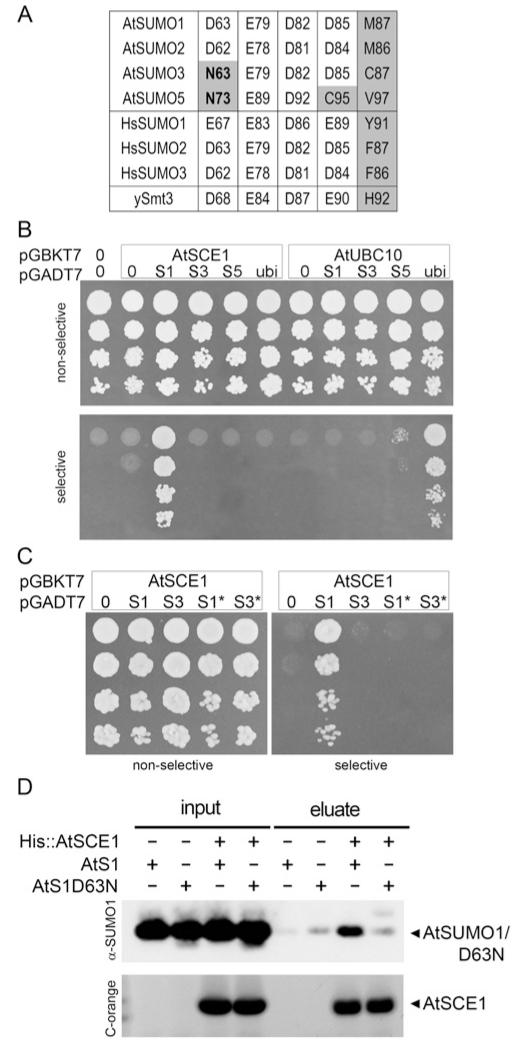
Figure 2. Non-covalent E2-interaction properties of AtSUMO isoforms.
(A) Sequence alignment of Arabidopsis (At), human (Hs) and yeast (y) SUMO residues involved in SUMO–E2 non-covalent interaction through their lateral chain contacts. Non-conserved residues are highlighted in grey, and residues exclusively non-conserved in AtSUMO3/5 are in bold. (B) Yeast two-hybrid assay to study interactions between AtSUMO1, 2, 3, 5 or ubiquitin and AtSCE1 or AtUBC10. (C) Interaction analysis between the mutant AtSUMO1/D63N (S1*) or AtSUMO3/N63D (S3*) and AtSCE1 as in (B). Native SUMO isoforms were also included as a control. (D) Poly-histidine pull-down assay of AtSUMO1 or the AtSUMO1/D63N mutant variant using His–AtSCE1 as a bait. Incubations in the absence of the bait or the prey were used as negative controls. Aliquots of input and eluate fractions were resolved by SDS/PAGE. AtSUMO1/D63N and AtSCE1 were analysed by immunoblotting or Coomassie Fluor Orange staining (C-orange) respectively.