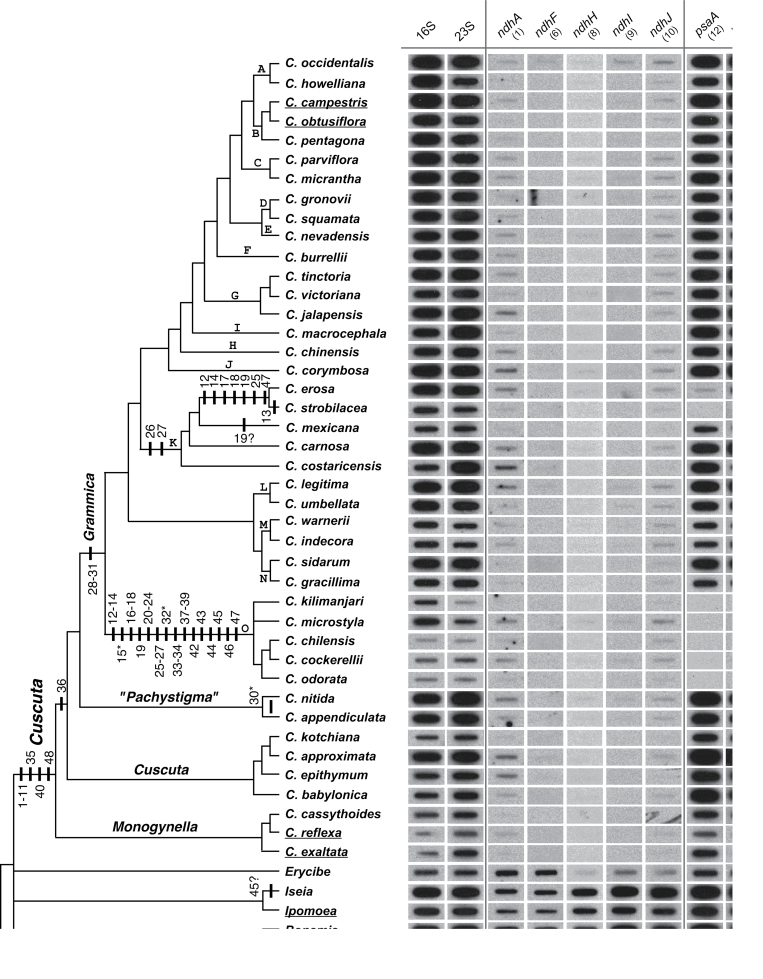
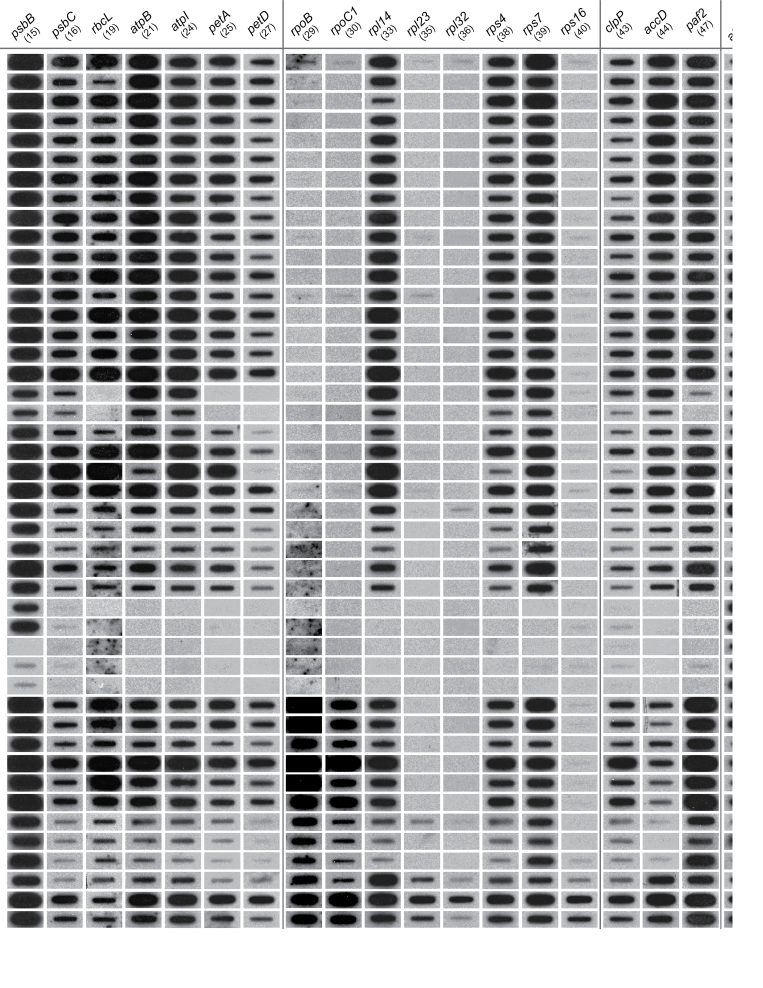
Fig. 1.
Autoradiographs representing a subset of slot-blot hybridization results for the presence/absence of 48 plastid protein-coding genes in Cuscuta and its close outgroups presented in a phylogenetic context. The topology shown is a composite tree depicting current understanding of relationships within Cuscuta derived from several published phylogenetic analyses (see text for references). Taxa with sequenced plastomes (Funk et al., 2007; McNeal et al., 2007a) are underlined. Parsimony reconstruction of plastid gene losses within Cuscuta under the assumption of irreversibility are mapped (bars) on the composite tree (depicted left). The numbers below each gene and above/below the bars refer to probes used in the hybridizations (see Supplmentary Table S1 at JXB online). Genes that are followed by a ‘?’ indicate potentially divergent copies of a plastid genes, whereas those followed by ‘*’ refer to genes that are present or absent (polymorphic) within the indicated clades. The plastid small (16S) and large (23S) rRNA subunits and mitochondrial ATP synthase subunit 1 (atp1) were used as positive controls (shown here is one representative out of five sets). Clades ‘K’ and ‘O’ show the greatest number of absences or near absences of hybridization signal for the plastid genes. Note that diminution of hybridization signal for clade ‘O’ extends to plastid positive controls (16S and 23S rDNA) compared with mitochondrial atp1. For full details, see Supplementary Table S1.