Fig. 4.
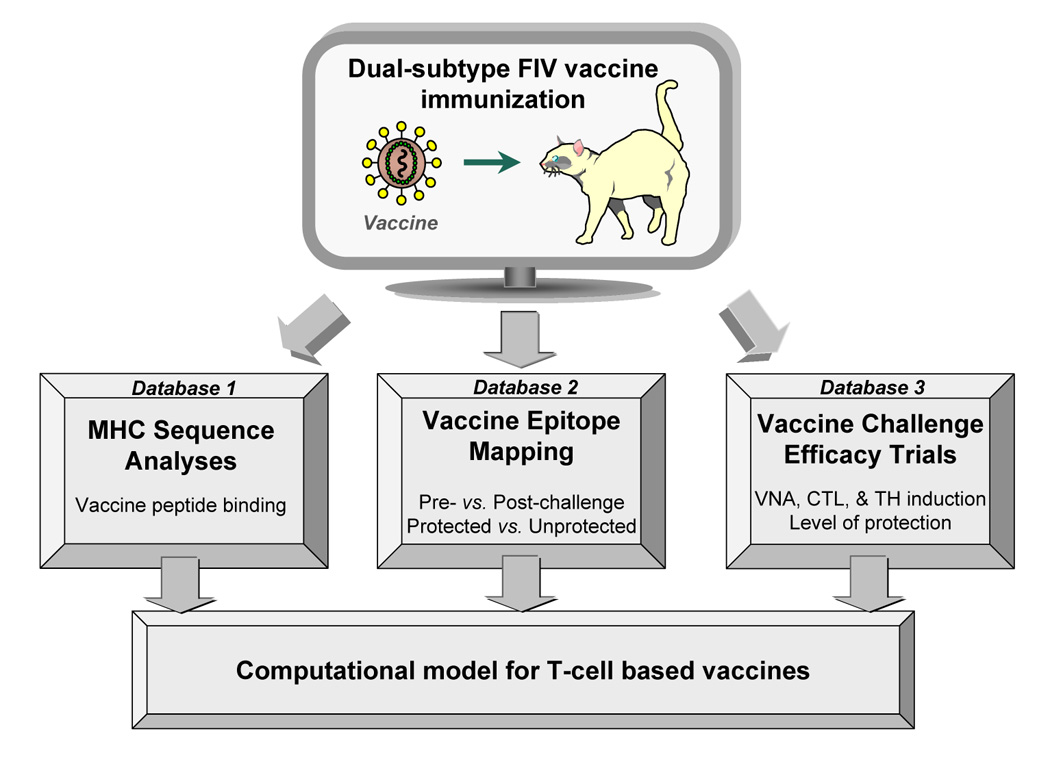
Computational modeling for T-cell immunity-based FIV vaccines. Propred-I and Propred-II epitope prediction tools are few of the known databases for identifying peptides binding to HLA class-I and class-II, respectively (De Groot et al., 2002). Los Alamos National Laboratory (LANL) database provides antibody, TH, and CTL epitope mapping for HIV-1 mainly derived from infected humans or vaccinated humans or animals (LANL, 2006). Similar epitope prediction tools based on feline MHC-I and –II can be produced concurrent to the sequencing of MHC alleles in domestic cat population (database 1) and to the epitope mapping generated by T-cell assays using semi-inbred cats (database 2). Vaccine epitope analysis can be performed directly in the natural host by immunizing potential protective peptides and evaluating the protective efficacy of these peptides against FIV challenge (database 3).
