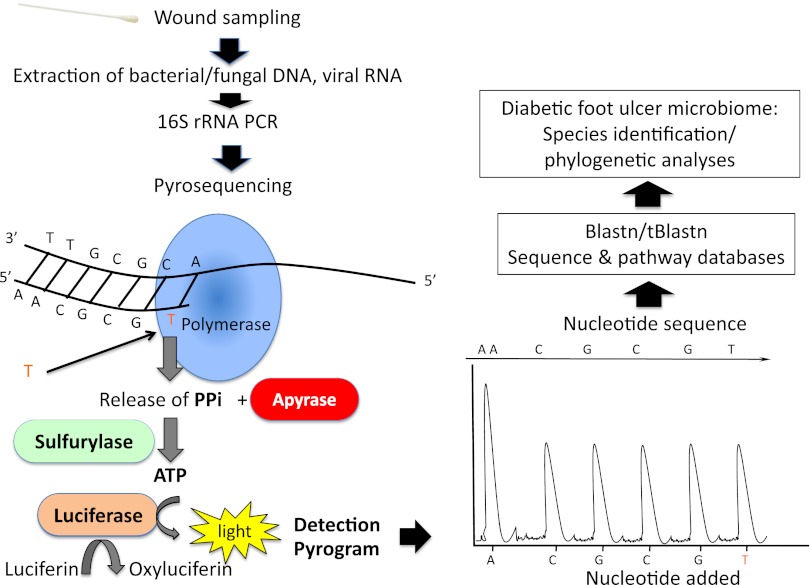
FIG. 1.
Steps for analyzing the microbiome of a DFU (adapted from England R, Pettersson M. Pyro Q-CpG: quantitative analysis of methylation in multiple CpG sites by Pyrosequencing. Nat Methods 2005;2:i–ii). After wound sampling, extraction is performed to obtain bacterial/fungal DNA and/or viral RNA. Specific genetic targets of hypervariable regions within bacterial 16S rRNA genes are amplified by the polymerase chain reaction (PCR) and subjected to DNA pyrosequencing. Sequencing by synthesis occurs by a DNA polymerase–driven generation of inorganic pyrophosphate (PPi) with the formation of ATP and ATP-dependent conversion of luciferin to oxyluciferin. The generation of oxyluciferin causes the emission of light pulses, and the amplitude of each signal is directly related to the presence of one or more nucleosides. The different bacteria can be identified by the effective combination of conserved primer-binding sites and intervening variable sequences that facilitate genus and species identification (software Blastn/tBlastn, http://blast.ncbi.nlm.nih.gov/). The identification of all the bacterial genomes amplified determines the composition of DFU flora (microbiome).