Abstract
The sigma-1 receptor is a ligand-regulated ER resident chaperone involved in the maintenance of cellular homeostasis. Coupling of the sigma-1 receptor with various ER and/or plasma membrane ion channels is associated with its ability to regulate the locomotor activity and cellular proliferation produced in response to sigma-1 receptor ligands. A number of endogenous small molecules bind to the sigma-1 receptor and have been shown to regulate its activity; these include progesterone, N,N-dimethyltryptamine, D-erythro-sphingosine, and/or other endogenous lipids. We previously reported the synthesis of long chain N-alkylamine derivatives and the characterization of the structural-activity relationship between chain length of N-alkylamine and affinities at the sigma-1 receptor. Here, we present data demonstrating the photo-incorporation of one of these N-alkylamine derivatives, 4-NPPC12, to the sigma-1 receptor. MALDI-TOF-TOF and tandem mass spectrometry showed that 4-NPPC12 photo-inserted at histidine 154 of the derivatized population of the sigma-1 receptor. Interestingly, light dependent photo-insertion of 4-NPPC12 resulted in an enhanced electrophoretic mobility of only 50% of the derivatized receptor molecules as assessed by SDS polyacrylamide gel electrophoresis. The proposed binding and reactivity of 4-NPPC12 evokes a ligand binding model for the sigma-1 receptor that likely involves a receptor dimer and/or oligomer.
Keywords: Sigma-1 receptor, dimer, photoaffinity labeling, sphingolipids, 4-nitrophenylpropyl, mass spectrometry, crosslinking, gel mobility shift
The sigma-1 receptor belongs to a unique family of membrane receptors of which there are at least two subtypes, the sigma-1 (26 kDa) and sigma-2 (18 kDa) receptors. The sigma-1 and sigma-2 receptors are distinguishable by their pharmacological profiles, selectivity, functions, subcellular locations, and molecular weight. Since its sequence is available (1), the sigma-1 receptor has been extensively evaluated and shown to possess chaperone activity (2). The 18 kDa sigma-2 receptor has not yet been cloned but recently a 26 kDa membrane localized progesterone receptor membrane component 1 (PCRMC1) has been shown to possess sigma-2 like binding site (3). Multiple lines of evidence indicate that the sigma-1 receptor plays an important role in the maintenance of cellular membrane potential via a direct and/or indirect modulation of various ion channels. These studies demonstrated that the sigma-1 receptor can regulate the function of voltage-gated potassium (K+) (4), sodium (Na+) (5), calcium (Ca2+) (6), chloride (Cl-) channels (7) and expression of the human hERG potassium channel (8). Direct physical interactions between the sigma-1 receptor and voltage-gated potassium channels (4) and between the sigma-1 receptor and the acid-sensing ion channel ASIC1a (9) have also been demonstrated leading to the suggestion that the sigma-1 receptor may be a regulatory subunit for certain ion channels. Additionally, through its chaperone activity, the sigma-1 receptor can indirectly modulate Ca2+ entry from the ER into the mitochondria via the stabilization of the inositol 1,4,5-trisphophate receptor type 3 (IP3R-3) (2, 10).
Interests in the sigma-1 receptor arise from its “promiscuous” interaction with a wide range of synthetic pharmacological compounds such as the antipsychotic, haloperidol; Ca2+ channel antagonist, verapamil; antidepressants, fluvoxamine; and CNS stimulant, methamphetamine (for a literature review on sigma-1 receptor ligands see Su et al. (11) and on the sigma-1 receptor pharmacophore see Glennon et al. (12), publications by Fontanilla et al. (9), and Chu et al. (13)). Endogenously, the sigma-1 receptor has been suggested to bind a number of compounds including neurosteroids, progesterone (14); the hallucinogen, N,N-dimethyltryptamine (DMT) (15); and/or sphingolipids such as D-erythro-sphingosine (16) and ceramides (17). Ligand binding to the sigma-1 receptor has the following cellular signaling consequences: modulation of the interaction between an ER-resident chaperone GRP78/BiP and sigma-1 receptor (2), regulation of the association of ankyrin B, IP3 receptor and sigma-1 receptor tri-complex (18), influencing the ion flux through different ion channels (4-8), re-distribution of the sigma-1 receptor to different cellular locals (17, 19, 20) and a reduction of cellular oxidative stress (21-23).
The sigma-1 receptor cloned from various species encodes a protein of 223 amino acids (1, 24, 25) that migrates on SDS polyacrylamide at 26 kDa. We have been involved in defining the ligand-binding region of the sigma-1 receptor using photoaffinity labels (26-28). For example, we showed previously that a high affinity sigma-1 selective photolabel [125I]-Iodoazidococaine ([125I]-IACoc) photolabeled aspartate 188 located on steroid binding domain-like (SBDL) II (26, 29), [125I]-Iodoazidofenpropimorph ([125I]-IAF) photolabeled both SBDLI and SBDLII (27) and a radioiodinated benzophenone bifunctional crosslinking reagent tethered together SBDLI and SBDLII indicating an approximate 8 Å distance between these two domains (30). Additionally, C-terminal truncations of the last 15 amino acids from the C-terminal region of the sigma-1 receptor showed reduced [125I]-IACoc photolabeling (26). A number of biochemical studies from our laboratories as well as others using site directed mutagenesis have also demonstrated the importance of the C-terminal half of the sigma-1 receptor for ligand binding (26, 28, 31). For example, Ganapathy et al. (32) has shown that a sigma-1 receptor splice variant in Jurkat human T lymphocyte cells lacking amino acids 119-149 had reduced [3H]-haloperidol binding. Specifically, substitutions of aspartate 126 and glutamate 172 with glycine completely abolished [3H]-haloperidol binding to the sigma-1 receptor (31) in partial support for the idea that the C-term is important for ligand binding. In expanding our previous work on the binding of N-(3-phenylpropyl)alkan-1-amines and N-(3-(4-nitrophenyl)propyl)alkan-1-amines (PPC4, PPC7, PPC12, PPC18, 4-NPPC4, 4-NPPC7, 4-NPPC12, and 4-NPPC18) (13) to the sigma-1 receptor, here we report a high yield covalent photo-incorporation of one of these molecules, 4-NPPC12. The resulting photo-crosslinking of 4-NPPC12 to the sigma-1 receptor resulted in an increase in the electrophoretic mobility (by 3 kDa) of half the population of the sigma-1 receptor molecules. MALDI-TOF-TOF and tandem mass spectrometry identified histidine 154 as the site of 4-NPPC12 covalent photo-incorporation. Mechanistically, the previously proposed “half site” reactivity of [3H]-(+)-pentazocine binding to the pure sigma-1 receptor suggests that 4-NPPC12 may bind to a dimer of the receptor with a stoichiometry of 2 to 1 receptor monomer to ligand ratio.
Experimental procedures
Reagents and constructs
[3H]-(+)-Pentazocine and [3H]-(+)-1,3-ditolyl guanidine (DTG) were obtained from Perkin Elmer, Waltham, MA. N-3-phenylpropyl-N-alkylamines and N-3-(4-nitrophenyl)propyl-N-alkylamines were synthesized previously (13). The guinea pig maltose binding protein sigma-1 receptor fusion was created previously (33). All sigma-1 receptor mutants including the histidine 154 alanine (H154A), cysteine 94 alanine (C94A), and cysteine 94 alanine / methionine 170 cysteine (C94A/M170C) double mutants were created in the pcDNA3.1 plasmid using site directed mutagenesis (Invitrogen, Carlsbad, CA). The anti-sigma-1 receptor antibody was developed and characterized as described previously (33) while the sequence specific antibodies against amino acid 52 – 69 or 143 – 165 antibodies (2) were donated by Dr. T. Hayashi and Dr. TP Su, NIDA.
Expression and purification of the sigma-1 receptor from E. coli
The procedure for the purification of the guinea pig sigma-1 receptor was described previously (33). Briefly, E. coli strain BL21(DE3) (Novagen, Madison, WI) containing the maltose-binding protein-sigma-1 receptor-6-histidine were grown to an OD600 of 0.6 before induction with 0.5 mM IPTG for 4 h at 37°C. Cells were collected by centrifugation and the E. coli pellet was resuspended in buffer I (20 mM Tris-Cl pH 7.5, 200 mM NaCl, 1 mM 2-mercaptoethanol, and 1 mM EDTA). The cell suspension was sonicated using a Branson soniWer 250 employing a 1 cm probe (output 50%, 2 s bursts, 1 s lag) for 15 min on ice. The cell lysate was centrifuged at 100,000 g for 1 h to separate total particulate and soluble proteins. The particulate fraction was extracted with Triton X-100 at a 4:1 ratio of detergent to total protein (w/w) for 3 h with gentle stirring at 4°C. The extracted material was centrifuged again at 100,000 g for 1 h and the supernatant was diluted with buffer I to obtain a Triton X-100 concentration of 1%. Proteins were loaded onto an amylose column (New England Biolabs, Ipswich, MA), washed once with 5 column volumes of buffer II (20 mM Tris-Cl pH 7.5, 200 mM NaCl, 1 mM 2-mercaptoethanol, 1 mM EDTA, 0.5% TX-100) and once with 3 column volumes of buffer III (20 mM Tris-Cl pH 7.5, 200 mM NaCl, 5 mM CaCl2, 0.5% TX-100). The MBP-sigma-1 receptor fusion protein was eluted with 3 column volumes of buffer IV (20 mM Tris-Cl pH 7.5, 200 mM NaCl, 5 mM CaCl2, 10 mM maltose, 0.5% TX-100).
The pure MBP-sigma-1-receptor fusion protein was cleaved with Factor Xa protease (Novagen, Madison, WI) in 5 ml fractions at RT for 24 – 48 h and the cleavage monitored by SDS-polyacrylamide gel electrophoresis. The sigma-1 receptor from the Factor Xa cleavage was purified with HIS-Select HC Nickel affinity gels (Sigma, St. Louis, MO) in a batch format. Proteins and Ni2+ beads slurry were tumbled overnight at 4°C, then washed 3 times with buffer V (50 mM Na2HPO4 pH 8, 200 mM NaCl, 0.5% TX-100), and eluted with buffer VI (50 mM Na2HPO4 pH 8, 200 mM NaCl, 250 mM imidazole, 0.5% TX-100) at RT.
Preparation of guinea pig liver membranes (GPLM) and rat liver membranes (RLM)
Membranes were prepared as described previously (29) from frozen tissues (Pel Freez Biologicals, Rogers, AR). The liver tissue was homogenized (10 ml buffer/g wet tissue) by 4 bursts of 10 s each using a brinkman polytron (American Laboratory Trading Inc., East Lyme, CT) on setting 6 in ice cold sodium phosphate buffer (10 mM pH 7.4) containing 0.32 M sucrose and a cocktail of protease inhibitors (20 μg/ml leupeptin, 5 μg/ml soybean trypsin inhibitor, 100 μM phenylmethylsulfonyl fluoride (PMSF), 100 μM benzamidine and 1 mM EDTA). The membrane suspension after homogenization was centrifuged for 10 min at 17,000 g and the supernatant was further centrifuged at 100,000 g to collect the membrane fraction. The pellet from the 100,000 g centrifugation was resuspended in the same buffer as above, snap frozen and stored at -80°C at a protein concentration of 10 mg/ml.
Transient expression of the sigma-1 receptor in COS-7 cells
The guinea pig sigma-1 receptor in pcDNA3.1 was transfected into COS-7 cells by electroporation and grown for 48 hr before harvested with trypsin. Cells were then resuspended in 1.5 ml of 1X PBS containing protease inhibitor cocktail (Sigma-Aldrich, St. Louis, MO) and homogenized by passaging through a 27-gauge syringe 25 times. Protein concentrations were determined by the Bio-Rad Protein Assay reagent (Bio-Rad, Hercules, CA).
Photolabeling and western analyses
Fifty micrograms of guinea pig liver membranes (GPLM) or lysates from COS-7 cells overexpressing the sigma-1 receptor were incubated with 10 μM of the test compounds for 30 min at RT. The reaction mixtures were then illuminated for 10 s with a high pressure AH6 mercury lamp to activate the photoprobe followed by separation on a 12% SDS polyacrylamide gel. Proteins were transferred to a polyvinyldifluoride (PVDF) membrane (Millipore, 0.45 μm) in 10 mM 3-(Cyclohexylamino)-1-propanesulfonic acid (CAPS) pH 10.5 containing 0.5 % w/v DTT and 15 % v/v methanol at 65 V for 1 h at 4°C. The PVDF membrane was blocked with 5% non-fat dry milk for 1 h at RT, probed with anti-sigma-1 receptor overnight at 4°C, washed 3 times for 10 min each in PBS followed by incubation with secondary anti-Rabbit IgG HRP for 1 h at RT. The membrane was then washed thoroughly and developed with enhanced chemiluminescence (ECL) reagents (Pierce, Rockford, IL). Protection experiments were performed by pre-treating the protein samples with various sigma-1 receptor ligands prior to the addition of 4-NPPC12. Western analyses of the sigma-1 receptor with sequence specific antibodies were performed similarly.
Endoproteinase LysC (EndoLysC) digestion
EndoLysC cleavage of the photolabeled sigma-1 receptor was performed according to Pal et al. (27). The E.coli expressed sigma-1 receptor was first labeled with 4-NPPC12 followed by separation on a 12% SDS gel. The S1R26kDa and the S1R23kDa bands were separately excised from the gel, macerated, and eluted with 1 ml of water by tumbling overnight at RT. Supernatant containing the sigma-1 receptor was collected and concentrated to 100 μL by lyophilization and treated with EndoLysC (0.25 μg) overnight at RT. The digestion was terminated by adding 5X SDS-PAGE sample buffer, and samples were resolved on a 16.5% SDS-Tricine/PAGE. The protein samples were transferred to a PVDF membrane for western analyses as described above using the sequence specific antibodies against amino acid 52 – 69 or 143 – 165 antibodies, the polyclonal sigma-1 receptor developed in our laboratory (33), or the C-terminal hexahistidine tag antibody (Invitrogen, Carlsbad, CA).
“In Gel” digestion with trypsin and mass spectrometry determination
“In Gel” digestion and mass spectrometric analyses were performed at the Mass Spectrometry Facility (Biotechnology Center, University of Wisconsin-Madison). For detailed methodologies refer to the Supplemental Information section.
Saturation binding of [3H]-(+)-pentazocine and [3H]-DTG
Saturation binding of [3H]-(+)-pentazocine to the sigma-1 receptor WT and the histidine 154 alanine mutant (H154A) or [3H]-DTG to the cysteine 94 alanine (C94A) and cysteine 94 alanine / methionine 170 cysteine (C94A/M170C) were carried out in a total volume of 100 μL containing 30 μg of the total cell lysates from COS-7 cells overexpressing each mutant in 50 mM Tris-Cl, pH 8 and concentrations of [3H]-(+)-pentazocine from 1 nM – 100 nM for 60 min at 30°C. The reaction was terminated by rapid filtration through glass fiber filters (Whatman GF/B, Whatman, Maidstone, UK), using a Brandel cell harvester (Brandel, Gaithersburg, MD). The glass fiber filters were pre-soaked in 0.5% polyethyleneimine (PEI) for at least 1 h at RT. Filters were washed 4 times with 4 ml of ice-cold 50 mM Tris-Cl, pH 8.0. Radioactivity was quantified by liquid scintillation (Ultima Gold, Perkin Elmer, Waltham, MA) counting using a liquid scintillation counter (Packard model 1600CA, Packard Instrument Co., Downers Grove, IL).
Competitive displacement of [3H]-(+)-pentazocine binding
The conditions for the competition displacement of [3H]-(+)-pentazocine to the WT and H154A sigma-1 receptors were performed similar to saturation binding except all samples contained a final concentration of 10 nM [3H]-(+)-pentazocine and increasing concentrations of 4-NPPC12 between 3 nM and 100 μM. IC50 values were then converted to KI values using the Cheng – Prusoff correction (34) with the following equation KI = IC50/(1 + [L]/KD) where [L] is the ligand concentration, and KD is the previously determined dissociation constant for [3H]-(+)-pentazocine (10 nM).
Results
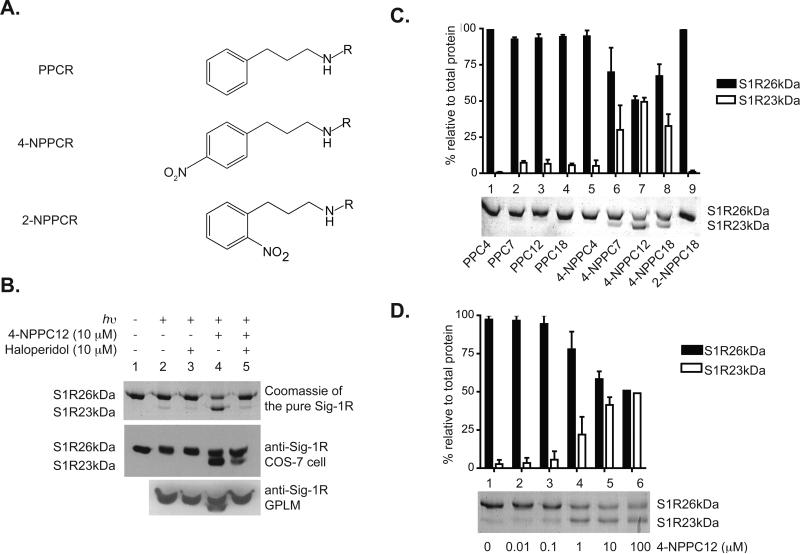
Previously, we reported the synthesis and evaluation of N-(3-phenylpropyl) and N-(3-(4-nitrophenyl)propyl) derivatives of N-alkylamine with varying chain length (Figure 1A) as high affinity ligands for sigma-1 and sigma-2 receptors (13). Binding studies showed that affinities increased linearly with carbon chain length and maximum binding was achieved with 12 carbon atoms. Additionally, four of these N-alkylamine derivatives (PPC7, 4-NPPC7, PPC12, and 4-NPPC12) showed high selectivity for the sigma-1 and sigma-2 receptors when assessed in a binding screen of over 40 other membrane receptor/transporter targets and these four compounds have demonstrated promising anti-cancer activities against an array of cancer cell lines. At present, we report the discovery of photo-crosslinking by one of these molecules, 4-NPPC12, to the sigma-1 receptor. The unique photochemistry of 4-NPPC12 allowed its covalent photo-incorporation into the sigma-1 receptor and resulted in an alteration of the electrophoretic migration pattern of the receptor on SDS gels. Illumination using a high-energy mercury lamp (hν) in the presence of an equal molar ratio of the pure sigma-1 receptor and 4-NPPC12 produced a secondary species, the S1R23kDa form (Figure 1B). The secondary species was named for its apparent molecular weight of 23kDa on SDS polyacrylamide gels, approximately 3 kDa smaller than the sigma-1 receptor apparent gel size of 26 kDa (Figure 1B). Furthermore, we found that while light alone produced less than 10% of the S1R23kDa species (Figure 1B, lane 2), 4-NPPC12 in combination with light maximized the production of the S1R23kDa form up to 50% of total protein used in each reaction (Figure 1B, lane 4). We could also detect the S1R23kDa species in both COS-7 cells overexpressing the guinea pig sigma-1 receptor and in guinea pig liver membranes (Figure 1B, middle and lower) using a full-length sigma-1 receptor polyclonal antibody (33). The generation of the S1R23kDa form from both membrane preparations confirmed that the pure sigma-1 receptor is structurally similar to the receptor in situ, justifying our use of the pure sigma-1 receptor in subsequent studies. Since the gel migration pattern of the S1R23kDa species could have been due to a non-specific photochemical reaction of 4-NPPC12 to the sigma-1 receptor, we found that pre-incubation with a putative sigma-1 receptor antagonist, haloperidol and DTG, could block the formation of the S1R23kDa form (Figure 1B, lane 5 and Supplementary Figure S2C) indicating specific photo-crosslinking at (or near) the haloperidol binding site. Moreover, we were unable to detect the S1R23kDa species when the pure protein was pre-denatured with 1% SDS (data not shown).
Figure 1.
Asymmetric photo-crosslinking of 4-NPPC12 to the sigma-1 receptor. A. Chemical structures of N-alkylamine derivatives: PPC4 and 4-NPPC4, R = nButyl; PPC7 and 4-NPPC7, R = nHeptyl; PPC12 and 4-NPPC12, R = nDodecyl; PPC18, 4-NPPC18 and 2-NPPC18, R = nOctadecyl. These compounds were previously reported (13). B. Light- and 4-NPPC12-dependent formation of the sigma-1 receptor 23kDa were observed in different preparations of the sigma-1 receptor: coomassie stained gel of the pure sigma-1 receptor (upper), lysates from COS-7 cells overexpressing the sigma-1 receptor – detected via western blots (middle), and guinea pig liver microsomes (GPLM) – also detected via western blotting (lower). C. The formation of the lower 23 kDa species was determined to be dependent on alkyl chain length and the presence of a 4-nitro moiety. A representative Coomassie stained gel of the pure sigma-1 receptor treated with varying chain length N-alkylamine derivatives in the presence of light. D. The sigma-1 receptor 23 kDa was also dependent on the concentrations of compound, 4-NPPC12, as shown from Coomassie staining of the pure sigma-1 receptor. Each experiment was performed three times (error bar = SEM).
We next explored the structural requirements of N-alkylamine derivatives leading to the formation the S1R23kDa species. Since a number of N-alkylamine derivatives were available (Figure 1A) (13), we first determined the contribution of the nitro moiety and/or carbon chain length of N-alkylamine derivatives on the generation of the S1R23kDa species. Non-nitro (PPC4, PPC7, PPC12, and PPC18) and 2-nitro (2-NPPC18, KI value of 76 ± 3 nM at the pure sigma-1 receptor) molecules failed to produce the S1R23kDa species (Figure 1C, lanes 1 – 4) while three of the 4-nitro molecules (4-NPPC7, 4-NPPC12, and 4-NPPC18) all generated the S1R23kDa form (Figure 1C, lanes 6 – 8). Interestingly, the N-butylamine nitro derivative, 4-NPPC4, was also unable to produce the S1R23kDa species (Figure 1C, lane 5) but compounds with at least seven carbons all produced this secondary sigma-1 receptor form. While the mobility shift pattern of the S1R23kDa form suggested an affinity dependency, 4-NPPC7, with an affinity of 7.5 nM, was still less efficient than 4-NPPC12 (KI value of 32 nM) at producing the secondary species. These results indicated that the photo-crosslinking of 4-NPPC12 to the sigma-1 receptor is carbon chain length dependent and that the dodecylamine nitro derivative, 4-NPPC12, is optimally situated to react with the sigma-1 receptor within the binding pocket. Next, we examined the dependence of ligand concentration for the formation of the secondary S1R23kDa species. First, we found that treatment with 4-NPPC12 up to 100 μM, 10 fold above the concentration of receptors, did not enhance the production of the S1R23kDa form beyond 50% (Figure 1D). Second, the formation of the S1R23kDa species was independent of the time of light exposure since multiples of 10 s light bursts and extended light exposure up to 60 s (data not shown) did not enhance the formation of the S1R23kDa species. The generation of the S1R23kDa form was obtained maximally at an appropriate 1:1 molar ratio of the S1R26kDa to the S1R23kDa forms in these experiments.
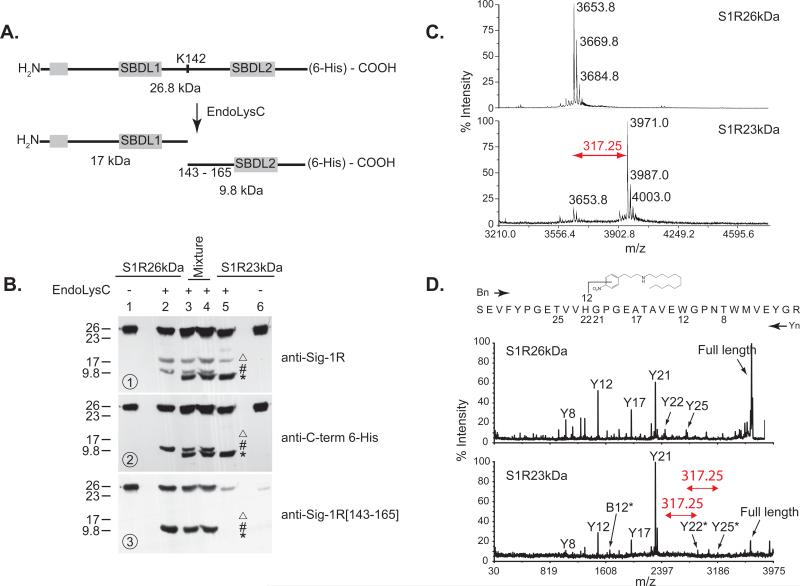
In an attempt to characterize the differences between the S1R26kDa and the S1R23kDa form, we took two approaches: (1) EndoLysC cleavage of both forms of the receptors followed by detection of the peptide fragments with sequence specific antibodies and (2) MALDI-TOF-TOF analysis of peptides generated from trypsin digestion of both forms. Figure 2A shows the linear representation of the sigma-1 receptor and its EndoLysC generated peptides (17 kDa and 9.8 kDa). Western analyses using a C-terminal hexahistidine antibody indicated that the 9.8 kDa peptide produced from the S1R23kDa species migrated lower than that of the 9.8 kDa equivalent from the S1R26kDa species (Figure 2B, second western). Furthermore, we found that while the sequence specific antibody against amino acid 143 – 165 recognized the 9.8 kDa peptide from the S1R26kDa form (Figure 2B, third western, lanes 2 - 4), this same antibody failed to detect the 9.8 kDa peptide generated from the S1R23kDa species (Figure 2B, third western, lanes 3 - 5). The discovery that the [143 – 165] sequence specific antibody failed to detect its epitope on the S1R23kDa species suggested that 4-NPPC12 covalently derivatized the sigma-1 receptor in this region.
Figure 2.
Identification of the 4-NPPC12 binding region on the pure sigma-1 receptor. A. Linear sequence representation of the guinea pig sigma-1 receptor and its endoproteinase lys C (EndoLysC) cleavage patterns. SBDL1, amino acids 91-109; SBDL2, amino acids 176-194. B. The gel mobility shift of the lower 23 kDa species was mapped to the C-terminal region of the sigma-1 receptor. Shown are western blots of peptides generated from the EndoLysC cleavage of the S1R26kDa and S1R23kDa forms with various sequence specific antibodies. Antibody against the full-length sigma-1 receptor (Sig-1R Ab), sequence specific antibodies against amino acids 143-165 ([143-165]Sig 1R Ab), and antibodies against the C-term 6-histidine tag (C-term 6-His Ab). 17 kDa, [Δ]; 9.8 kDa from the S1R26kDa species, [#]; and 9.8 kDa peptide from the S1R23kDa species, [*]. C. MALDI-TOF-TOF analyses localized 4-NPPC12 derivative to peptide region 143 – 175. Shown are selected spectral region of both S1R26kDa and S1R23kDa forms. 4-NPPC12 photolysis resulted in a mass addition of 317.25 Da to the tryptic peptide corresponding to amino acids 143-175 generated from the S1R23kDa form (lower) but not the S1R26kDa form (upper). For the complete MALDI-TOF-TOF results, see supplementary Figure S1. D. MS/MS mapping identified histidine 154 as the site of 4-NPPC12 photo-crosslinking to the lower S1R23 kDa form of the sigma-1 receptor. Shown are MS/MS spectra of the 143 – 175 peptides from both the underivatized S1R26kDa (upper) and the derivatized S1R23kDa forms (lower). Yn designates peptides generated from the C-terminus toward N-terminus and Bn are peptides generated from the N-terminus toward the C-terminus of the parent 143-175 tryptic peptide where n is the amino acid position in either direction. [*] denotes peptides that were shifted by 317.25 Da.
We therefore used MALDI-TOF-TOF analysis as a second approach to deduce the region of 4-NPPC12 photo-incorporation to the S1R23kDa form. Trypsin protease was chosen since the guinea pig sigma-1 receptor contains two lysines and several arginines and tryptic peptides generated from both the S1R26kDa and the S1R23kDa forms would also be within the detection limits of the MALDI-TOF-TOF analyses. While MALDI-TOF-TOF provided an overall sequence coverage of 55%, the C-terminal region of interest, amino acid residues 120-223, which were predicted to contain the reaction product of 4-NPPC12 was almost completely sequence covered (see supplementary Figures S1 and Tables S1 and S2 for detail results from MALDI-TOF-TOF experiments). MALDI-TOF-TOF spectral regions between 3210 and 4594 Da of the S1R26kDa and the S1R23kDa species were selected to highlight the differences between these two forms of the sigma-1 receptor created by the reaction with 4-NPPC12 (Figure 2C). For a complete MALDI-TOF-TOF spectra of both the S1R26kDa and S1R23kDa species, refer to Supplementary Figure S1. A tryptic peptide corresponding to amino acids 143-175 from the S1R26kDa species produced a molecular mass of 3653.8 Da (3669.8 Da represents the single methionine oxidized form, and 3684.8 Da represents the double methionine oxidized form), closely matched the predicted size. However, the same peptide from the S1R23kDa species produced a molecular mass of 3971.0 Da (and 3987.0 Da represents the single methionine oxidized form, and 4003.0 represents the double methionine oxidized form), 317.25 Da higher than its equivalent peptide generated from trypsin digestion of the S1R26kDa form.
Tandem MS/MS of the modified 143-175 peptide from the S1R23kDa species showed that amino acid histidine 154 contained a 317.25 Da adduct (Figure 2D and Tables S3 and S4). As summarized in Figure 2D (and Tables S3 and S4), while the observed mass of peptide Y21 [GPGEATAVEWGPNTWMVEYGR] matched its theoretical mass, Y22 [HGPGEATAVEWGPNTWMVEYGR] which contains an additional histidine, showed an increase in 317.25 Da. Likewise, peptide B12 [SEVFYPGETVVH] was also modified by an addition of 317.25 Da. Tryptic peptide 143-175 from the S1R26kDa species was treated similarly and showed the absence of the 317.25 Da addition (Figure 2D and Table S3) which validated the MALDI-TOF-TOF results. Taken together, these results indicated that 4-NPPC12 covalently modified the S1R23kDa form at histidine 154. While the 317.25 Da was 31.1 Da smaller than the molecular mass of 4-NPPC12 (348.25 Da), the adduct would be consistent with a loss of NOH from the phenyl ring region.
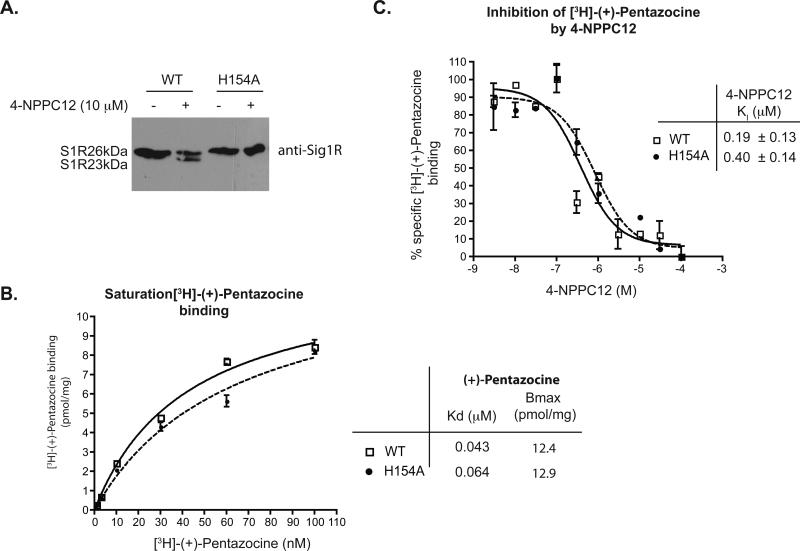
To confirm that histidine 154 was the site of 4-NPPC12 photo-incorporation, we substituted histidine 154 with alanine by site-directed mutagenesis. As shown in Figure 3A, while photolysis with 4-NPPC12 produced the S1R23kDa species in membranes of COS-7 cells overexpressing the WT sigma-1 receptor, the removal of histidine 154 abrogated the ability of this compound to produce the S1R23kDa form. Importantly, the failure of 4-NPPC12 to produce the S1R23kDa form was not due to an alteration in the ligand binding affinity since both [3H]-(+)-pentazocine (Figure 3B) and specifically 4-NPPC12 (Figure 3C) bound equally well to the H154A mutant as to the WT receptor. These results indicate that histidine 154 is situated in or close to the ligand binding site to allow a photochemical reaction with 4-NPPC12 to occur, but does not necessarily participate directly in the binding of ligand. Together, these data supported the conclusion that histidine 154 is the site of photo-derivatization by 4-NPPC12.
Figure 3.
H154A mutant binds equally efficient to [3H]-(+)-pentazocine and 4-NPPC12 but failed to form the S1R23kDa species upon photolysis. A. Mutation of the histidine 154 to alanine of the guinea pig sigma-1 receptor resulted in a failure of this mutant to form a light- and 4-NPPC12 dependent formation of the lower 23 kDa species. Shown is a western blot of cell lysates from COS-7 transfected with either the WT or H154A mutant and photolyzed with 4-NPPC12. B. The sigma-1 receptor H154A mutant retains binding to pentazocine. Saturationbinding of [3H]-(+)-pentazocine with lysates of COS-7 cell transfected with either WT or H154A mutant. C. The sigma-1 receptor H154A mutant binds to 4-NPPC12 similarly to the wildtype receptor. Inhibition of [3H]-(+)-pentazocine binding to determine the KI values for 4-NPPC12 for the WT sigma-1 receptors and the H154A mutant. All binding experiments were performed three times each in triplicates (error bar = SEM of the average).
Finally, it is important to note that the 317.2 Dalton adduct formed at histidine 154 is 31 Dalton smaller than the molecular mass of 4-NPPC12 (348.2 Dalton). Possibilities to account for the loss of 31 Dalton include (1) a loss of 2 oxygens with an addition of a proton, or (2) a loss of a nitrogen, an oxygen and a proton. A possible mechanism for covalent modification of histidine 154 by 4-NPPC12 may involve a photochemical rearrangement of 4-NPPC12 at the alpha carbon of the alkyl side chain to produce a double bond Michael acceptor. This would promote reaction with one of the amino groups of the imidazole ring (2-NPPC12, on the other hand, is likely to photochemically rearrange so that the proximal nitro group reacts intramolecularly thus preventing a reaction of the imidazole ring histidine 154). Detailed characterization of the light-dependent reaction mechanism between histidine and 4-NPPC12 is beyond the scope of the work reported here but future studies aiming at the elucidation of the photochemistry of the reaction of 4-nitro phenylpropyl compounds with imidazoles is warranted.
Discussion
Interest in the sigma-1 receptor has mainly revolved around its promiscuity in ligand binding to a wide range of pharmacologically important molecules. Determination of the ligand binding site therefore should assist in future designs of more selective ligands targeting the sigma-1 receptor as potential therapeutics against cancer, neurodegeneration, and psychosis. Previously, we synthesized and evaluated the binding characteristics of varying chain length N-alkylamines and their derivatives for the sigma-1 receptor (13). In studying the ligand binding region of N-alkylamine derivatives for the sigma-1 receptor, we uncovered a unique photo dependent property of three N-(3-(4-nitrophenylpropyl)alkan-1-amines (4-NPPC7, 4-NPPC12, and 4-NPPC18). Photolysis of 4-NPPC7, 4-NPPC12, or 4-NPPC18 with the sigma-1 receptor produced a novel secondary sigma-1 receptor form that migrated at 23 kDa (Figure 1A) – approximately 3 kDa smaller than its apparent migration patterns on SDS gels. From MALDI-TOF-TOF and MS/MS, 4-NPPC12 was found to be covalently derivatized to the S1R23kDa form at histidine 154 (Figures 2C and D), which partly explains the gel mobility shift of the full length S1R23kDa form and its EndoLysC-generated 9.8 kDa peptide (Figure 2B). Anomalous SDS-PAGE migration of some membrane proteins has been demonstrated to result from altered SDS binding related to the secondary structures of the proteins (35). Interestingly, the conjugation of a rat microtubule-associated light chain 3 (LC3) by phosphatidylethanolamine (PE), a marker of autophagy, also showed a downward-shift in gel mobility by 3 kDa (36). While it is generally believed that an increase in hydrophobicity (i.e. PE conjugation to LC3 or the derivatization of 4-NPPC12 to the S1R23kDa species) allowed more SDS loading thus retarding the electrophoretic mobility of proteins, in both cases the modified proteins migrated faster than their native forms. Hence, it may be a general phenomenon that lipidated forms of some proteins migrate faster on SDS-PAGE by as yet unknown mechanisms.
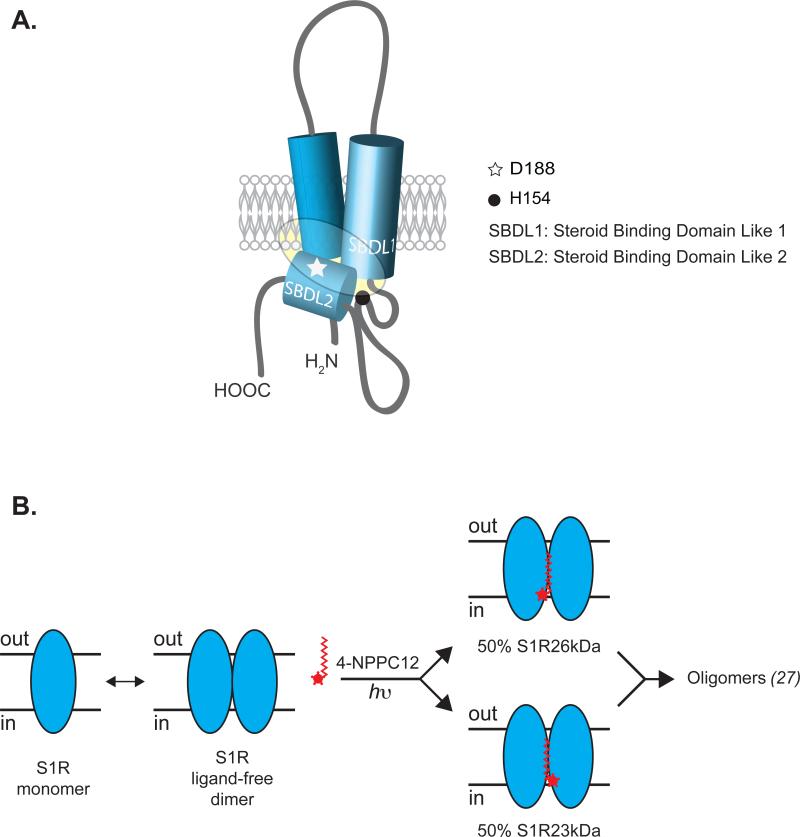
MS/MS results indicated that 4-NPPC12 formed a 317.25 Da adduct at histidine 154 of the S1R23kDa species. Substitution of histidine 154 with alanine, while preventing the formation of the S1R23kDa species induced by 4-NPPC12, did not alter binding of 4-NPPC12 and/or [3H]-(+)-pentazocine (Figure 3). We showed previously that SBDLI, SBDLII and part of the TM1 region form the binding site of sigma-1 receptor through the use of a number of photoaffinity labels (26-28). One of these photoprobes, [125I]-IACoc, was found to specifically photo-insert at aspartate 188 (26). The covalent crosslinking of 4-NPPC12 to histidine 154 has prompted new ideas regarding the sigma-1 receptor ligand-binding region. In the model presented in Figure 4A, the loop region between SBDLI and SBDLII is proposed to fold back to create a ligand binding pocket together with previously defined regions (Figure 4A, shaded area). Ganapathy et al. (32) demonstrated that a sigma-1 receptor splice variant in Jurkat human T lymphocyte cells lacking amino acids 119-149 was unable to bind to [3H]-haloperidol. We expressed this splice variant in COS-7 cells and found that photo-crosslinking with 4-NPPC12 did not produce the S1R23kDa form (data not shown) indicating the structural importance of this region for ligand binding to the sigma-1 receptor. Mutations of aspartate and glutamate 172 to glycine completely abolished [3H]-haloperidol binding (31) suggesting that the linker sequence between SBDLI and SBDLII is also important for ligand binding.
Figure 4.
Models of the sigma-1 receptor ligand binding region. A. A model of the sigma-1 receptor binding region from previous photolabeling studies and from the results presented here with the derivatization by 4-NPPC12. D188, aspartate 188; H154, Histidine 154; shaded area represents the ligand binding region. B. A proposed model of the sigma-1 receptor dimer in the presence of 4-NPPC12. The sigma-1 receptor may exist naturally in equilibrium between monomeric, dimeric, and/or oligomeric forms.
Another important observation from this work was the stoichiometric generation of the S1R23kDa form with 4-NPPC12. The light dependent 4-NPPC12-induced formation of the S1R23kDa was produced maximally at a 1 to 1 ratio of the S1R26kDa to the S1R23kDa forms. While the precise mechanism leading to this observation is unclear at the moment, we favor a hypothesis whereby the sigma-1 receptor forms a homodimer or an oligomer containing dimers in which 4-NPPC12 binds at the interface of the homodimer yet only photo-crosslinked stoichiometrically to one subunit of the dimer (Figure 4B). Multiple lines of evidence from our lab have suggested that the sigma-1 receptor may exist as a dimer or an oligomer in native membranes and in detergent solution. First, Pal et al. (27) found that both [125I]IACoc and [125I]IAF labeled oligomeric forms of sigma-1 receptors in rat and guinea pig liver membranes. Second, Ramachandran et al. (33) reported that the sigma-1 receptor expressed and purified from E.coli exhibited a 1:2 molar ratio [3H]-(+)-pentazocine binding to sigma-1 receptor respectively. Third, in COS-7 cells overexpressing the myc and HA epitope tagged sigma-1 receptor, co-immunoprecipitation using either one of the epitopes resulted in the pull down of the other epitope tagged sigma-1 receptor as evidence by western blotting (unpublished data). Finally, we have found here that upon mutating methionine 170 to cysteine in a receptor with a cysteine-less background, the M170C formed a SDS-resistant disulfide-linked dimer (as assessed by western blot using an anti-sigma-1 receptor antibody) which was eliminated by reducing agent(Supplementary Figure S2A). The sigma-1 receptor M170C mutant was found to possess similar binding affinity to the WT as assessed by saturating binding of [3H]-DTG (Supplementary Figure S2B). When considered collectively, our interpretation is that the sigma-1 receptor can naturally form dimers and/or oligomers. Together, these data evoke a model whereby the pure sigma-1 receptor exists as a homodimer and/or an oligomer containing dimers and binds to ligand with a ratio of one ligand (in this case 4-NPPC12) per dimer. We propose the following model to explain these results: 4-NPPC12 binds to the interface between two monomer subunits but reacts once with equal efficiency to either subunit which produces a 1 to 1 ratio of the unmodified form (S1R26kDa) to the modified form (S1R23kDa) of the sigma-1 receptor (Figure 4B). Alternatively, a second possibility could exist in which the sigma-1 receptor forms a structurally asymmetric homodimer and 4-NPPC12 preferentially reacts with only one subunit. A recent report on the structure of the abscisic acid (ABA) receptor (PYR1) in Arabidopsis thaliana showed that PYR1 forms an asymmetric homodimer in which only one monomer binds to ABA (37). The asymmetry of PYR1 is created by a cis-to-trans isomerization of a proline (Pro88) on the ligand bound monomer such that ABA is trapped in the ligand binding cavity. The isomerization also induced a conformational change of the protein domain at the interface of PYR1 homodimer allowing allosteric modulation by the isomerized subunit. Therefore, it is possible that the histidine 154 region is located at a dimerization interface and mediate conformational changes between homodimer subunits of the sigma-1 receptor within ligand binding. Additionally, the proposed interaction of 4-NPPC12 with the receptor may also be explained by the formation of higher order oligomers of the sigma-1 receptor with a dimer as the smallest functional unit (Figure 4B). Evidence for sigma-1 receptor dimerization is also supported in the protein sequence. We found that the sigma-1 receptor 1 receptor contains two GXXXG putative oligomerization motifs: the first one is located at TM2 (amino acid 87–91) and the second one in the region flanking the C-terminus of SBLDII (amino acids 108-112). These sequences have been shown to promote the formation of dimers in membrane bound glycophorin A (38) and oligomers in the membrane bound ABC transporter, ABCG2 (39). It may be possible that the first GXXXG sequence mediates the dimerization of the sigma-1 receptor while the second GXXXG sequence drives the formation of oligomers.
While the physiological significance of the sigma-1 receptor dimer in the context of the cellular environment is unclear, a recent report demonstrated that mutation of glutamic acid 102 to glutamine (E102Q) in the sigma-1 receptor produced a detergent resistant apparent homodimer as assessed by SDS PAGE (40). The sigma-1 receptor harboring an E102Q mutation was suggested by the authors to be causative for juvenile amyotrophic lateral sclerosis (JALS) in a consanguineous family with a high familial incidence of this neurodegenerative disease. Thus, it will be important in the future to determine the functional forms of the sigma-1 receptor in a cellular context, and to determine whether different classes of ligands (agonists versus antagonists) vary in their properties in binding to different quaternary forms of the receptor.
Supplementary Material
Acknowledgments
We wish to thank Dr. Teruo Hayashi and Dr. Tsung Ping Su (NIDA, NIH) for providing the sequence specific sigma-1 receptor antibodies, Dr. Michael Palte (Department of Biochemistry, University of Wisconsin – Madison) for insightful discussions of the possible photochemistry of 4-NPPC12 and 2-NPPC12, and Dr. Lian Wang Guo for reading this manuscript.
Research funding: This research was supported by National Institute of Health Grants MH065503 and GM-33138 (to A.E.R.), the Retina Research Foundation Edwin and Dorothy Gamewell Professorship (to A.E.R.), the UW-Madison Advanced Opportunity Fellowships (AOF) and the Gates Millennium Scholarship (to U.B.C).
ABBREVIATIONS
- DMT
N,N-dimethyltryptamine
- DTG
ditolylguanidine
- ER
endoplasmic reticulum
- GPLM
guinea pig liver membranes
- IP3R-3
inositol trisphophate receptor type 3
- MBP
maltose-binding protein
- SBDL
steroid-binding domain liked
- SKF-10,047
N-allyl-normetazocine
- TM
transmembrane
- RLM
rat liver membranes
Footnotes
Supplemental Information:
These data may be accessed free of charge via the internet at http://pubs.acs.org.
References
- 1.Hanner M, Moebius FF, Flandorfer A, Knaus HG, Striessnig J, Kempner E, Glossmann H. Purification, molecular cloning, and expression of the mammalian sigma1-binding site. P.N.A.S. 1996;93:8072–8077. doi: 10.1073/pnas.93.15.8072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hayashi T, Su TP. Sigma-1 receptor chaperones at the ER-mitochondrion interface regulate Ca(2+) signaling and cell survival. Cell. 2007;131:596–610. doi: 10.1016/j.cell.2007.08.036. [DOI] [PubMed] [Google Scholar]
- 3.Xu J, Zeng C, Chu W, Pan F, Rothfuss JM, Zhang F, Tu Z, Zhou D, Zeng D, Vangveravong S, Johnston F, Spitzer D, Chang KC, Hotchkiss RS, Hawkins WG, Wheeler KT, Mach RH. Identification of the PGRMC1 protein complex as the putative sigma-2 receptor binding site. Nat Commun. 2011;2:380. doi: 10.1038/ncomms1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aydar E, Palmer CP, Klyachko VA, Jackson MB. The sigma receptor as a ligand-regulated auxiliary potassium channel subunit. Neuron. 2002;34:399–410. doi: 10.1016/s0896-6273(02)00677-3. [DOI] [PubMed] [Google Scholar]
- 5.Johannessen M, Ramachandran S, Riemer L, Ramos-Serrano A, Ruoho AE, Jackson MB. Voltage-gated sodium channel modulation by sigma-receptors in cardiac myocytes and heterologous systems. American journal of physiology. Cell physiology. 2009;296:C1049–1057. doi: 10.1152/ajpcell.00431.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang H, Cuevas J. Sigma receptors inhibit high-voltage-activated calcium channels in rat sympathetic and parasympathetic neurons. J Neurophysiol. 2002;87:2867–2879. doi: 10.1152/jn.2002.87.6.2867. [DOI] [PubMed] [Google Scholar]
- 7.Renaudo A, L'Hoste S, Guizouarn H, Borgese F, Soriani O. Cancer cell cycle modulated by a functional coupling between sigma-1 receptors and Cl- channels. J Biol Chem. 2007;282:2259–2267. doi: 10.1074/jbc.M607915200. [DOI] [PubMed] [Google Scholar]
- 8.Crottes D, Martial S, Rapetti-Mauss R, Pisani DF, Loriol C, Pellissier B, Martin P, Chevet E, Borgese F, Soriani O. Sig1R protein regulates hERG channel expression through a post-translational mechanism in leukemic cells. The Journal of biological chemistry. 2011;286:27947–27958. doi: 10.1074/jbc.M111.226738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carnally SM, Johannessen M, Henderson RM, Jackson MB, Edwardson JM. Demonstration of a direct interaction between sigma-1 receptors and acid-sensing ion channels. Biophys J. 2010;98:1182–1191. doi: 10.1016/j.bpj.2009.12.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu Z, Bowen WD. Role of sigma-1 receptor C-terminal segment in inositol 1,4,5-trisphosphate receptor activation: constitutive enhancement of calcium signaling in MCF-7 tumor cells. J Biol Chem. 2008;283:28198–28215. doi: 10.1074/jbc.M802099200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Su TP, Hayashi T, Maurice T, Buch S, Ruoho AE. The sigma-1 receptor chaperone as an inter-organelle signaling modulator. Trends in pharmacological sciences. 2010;31:557–566. doi: 10.1016/j.tips.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glennon RA. Pharmacophore identification for sigma-1 (sigma1) receptor binding: application of the “deconstruction-reconstruction-elaboration” approach. Mini Rev Med Chem. 2005;5:927–940. doi: 10.2174/138955705774329519. [DOI] [PubMed] [Google Scholar]
- 13.Chu UB, Hajipour AR, Ramachandran S, Ruoho AE. Characterization of 4-Nitrophenylpropyl-N-alkylamine Interactions with Sigma Receptors. Biochemistry. 2011;50:7568–7578. doi: 10.1021/bi2004872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Su TP, London ED, Jaffe JH. Steroid binding at sigma receptors suggests a link between endocrine, nervous, and immune systems. Science. 1988;240:219–221. doi: 10.1126/science.2832949. [DOI] [PubMed] [Google Scholar]
- 15.Fontanilla D, Johannessen M, Hajipour AR, Cozzi NV, Jackson MB, Ruoho AE. The hallucinogen N,N-dimethyltryptamine (DMT) is an endogenous sigma-1 receptor regulator. Science. 2009;323:934–937. doi: 10.1126/science.1166127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramachandran S, Chu UB, Mavlyutov TA, Pal A, Pyne S, Ruoho AE. The sigma1 receptor interacts with N-alkyl amines and endogenous sphingolipids. European journal of pharmacology. 2009;609:19–26. doi: 10.1016/j.ejphar.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hayashi T, Fujimoto M. Detergent-resistant microdomains determine the localization of sigma-1 receptors to the endoplasmic reticulum-mitochondria junction. Mol Pharmacol. 2010;77:517–528. doi: 10.1124/mol.109.062539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hayashi T, Su TP. Regulating ankyrin dynamics: Roles of sigma-1 receptors. P.N.A.S. 2001;98:491–496. doi: 10.1073/pnas.98.2.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mavlyutov TA, Ruoho AE. Ligand-dependent localization and intracellular stability of sigma-1 receptors in CHO-K1 cells. Journal of molecular signaling. 2007;2:8. doi: 10.1186/1750-2187-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hayashi T, Su TP. Intracellular dynamics of sigma-1 receptors (sigma(1) binding sites) in NG108-15 cells. J Pharmacol Exp Ther. 2003;306:726–733. doi: 10.1124/jpet.103.051292. [DOI] [PubMed] [Google Scholar]
- 21.Pal A, Fontanilla D, Gopalakrishnan A, Chae YK, Markley JL, Ruoho AE. The sigma-1 receptor protects against cellular oxidative stress and activates antioxidant response elements. European journal of pharmacology. 2012;682:12–20. doi: 10.1016/j.ejphar.2012.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bucolo C, Drago F, Lin LR, Reddy VN. Sigma receptor ligands protect human retinal cells against oxidative stress. Neuroreport. 2006;17:287–291. doi: 10.1097/01.wnr.0000199469.21734.e1. [DOI] [PubMed] [Google Scholar]
- 23.Tuerxun T, Numakawa T, Adachi N, Kumamaru E, Kitazawa H, Kudo M, Kunugi H. SA4503, a sigma-1 receptor agonist, prevents cultured cortical neurons from oxidative stress-induced cell death via suppression of MAPK pathway activation and glutamate receptor expression. Neuroscience letters. 2010;469:303–308. doi: 10.1016/j.neulet.2009.12.013. [DOI] [PubMed] [Google Scholar]
- 24.Kekuda R, Prasad PD, Fei YJ, Leibach FH, Ganapathy V. Cloning and functional expression of the human type 1 sigma receptor (hSigmaR1) Biochem Biophys Res Commun. 1996;229:553–558. doi: 10.1006/bbrc.1996.1842. [DOI] [PubMed] [Google Scholar]
- 25.Seth P, Leibach FH, Ganapathy V. Cloning and structural analysis of the cDNA and the gene encoding the murine type 1 sigma receptor. Biochem Biophys Res Commun. 1997;241:535–540. doi: 10.1006/bbrc.1997.7840. [DOI] [PubMed] [Google Scholar]
- 26.Chen Y, Hajipour AR, Sievert MK, Arbabian M, Ruoho AE. Characterization of the cocaine binding site on the sigma-1 receptor. Biochemistry. 2007;46:3532–3542. doi: 10.1021/bi061727o. [DOI] [PubMed] [Google Scholar]
- 27.Pal A, Hajipour AR, Fontanilla D, Ramachandran S, Chu UB, Mavlyutov T, Ruoho AE. Identification of regions of the sigma-1 receptor ligand binding site using a novel photoprobe. Molecular Pharmacology. 2007;72:921–933. doi: 10.1124/mol.107.038307. [DOI] [PubMed] [Google Scholar]
- 28.Fontanilla D, Hajipour AR, Pal A, Chu UB, Arbabian M, Ruoho AE. Probing the steroid binding domain-like I (SBDLI) of the sigma-1 receptor binding site using N-substituted photoaffinity labels. Biochemistry. 2008;47:7205–7217. doi: 10.1021/bi800564j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kahoun JR, Ruoho AE. (125I)iodoazidococaine, a photoaffinity label for the haloperidol-sensitive sigma receptor. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:1393–1397. doi: 10.1073/pnas.89.4.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pal A, Chu UB, Ramachandran S, Grawoig D, Guo LW, Hajipour AR, Ruoho AE. Juxtaposition of the steroid binding domain-like I and II regions constitutes a ligand binding site in the sigma-1 receptor. The Journal of biological chemistry. 2008;283:19646–19656. doi: 10.1074/jbc.M802192200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seth P, Ganapathy ME, Conway SJ, Bridges CD, Smith SB, Casellas P, Ganapathy V. Expression pattern of the type 1 sigma receptor in the brain and identity of critical anionic amino acid residues in the ligand-binding domain of the receptor. Biochim Biophys Acta. 2001;1540:59–67. doi: 10.1016/s0167-4889(01)00117-3. [DOI] [PubMed] [Google Scholar]
- 32.Ganapathy ME, Prasad PD, Huang W, Seth P, Leibach FH, Ganapathy V. Molecular and ligand-binding characterization of the sigma-receptor in the Jurkat human T lymphocyte cell line. J Pharmacol Exp Ther. 1999;289:251–260. [PubMed] [Google Scholar]
- 33.Ramachandran S, Lu H, Prabhu U, Ruoho AE. Purification and characterization of the guinea pig sigma-1 receptor functionally expressed in Escherichia coli. Protein expression and purification. 2007;51:283–292. doi: 10.1016/j.pep.2006.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheng Y, Prusoff WH. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochemical pharmacology. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 35.Rath A, Glibowicka M, Nadeau VG, Chen G, Deber CM. Detergent binding explains anomalous SDS-PAGE migration of membrane proteins. P.N.A.S. 2009;106:1760–1765. doi: 10.1073/pnas.0813167106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. Embo J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nishimura N, Hitomi K, Arvai AS, Rambo RP, Hitomi C, Cutler SR, Schroeder JI, Getzoff ED. Structural mechanism of abscisic acid binding and signaling by dimeric PYR1. Science. 2009;326:1373–1379. doi: 10.1126/science.1181829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Overton MC, Chinault SL, Blumer KJ. Oligomerization, biogenesis, and signaling is promoted by a glycophorin A-like dimerization motif in transmembrane domain 1 of a yeast G protein-coupled receptor. J Biol Chem. 2003;278:49369–49377. doi: 10.1074/jbc.M308654200. [DOI] [PubMed] [Google Scholar]
- 39.Xu J, Peng H, Zhang JT. Human multidrug transporter ABCG2, a target for sensitizing drug resistance in cancer chemotherapy. Curr Med Chem. 2007;14:689–701. doi: 10.2174/092986707780059580. [DOI] [PubMed] [Google Scholar]
- 40.Al-Saif A, Al-Mohanna F, Bohlega S. A mutation in sigma-1 receptor causes juvenile amyotrophic lateral sclerosis. Ann Neurol. 2011;70:913–919. doi: 10.1002/ana.22534. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.