FIGURE 5.
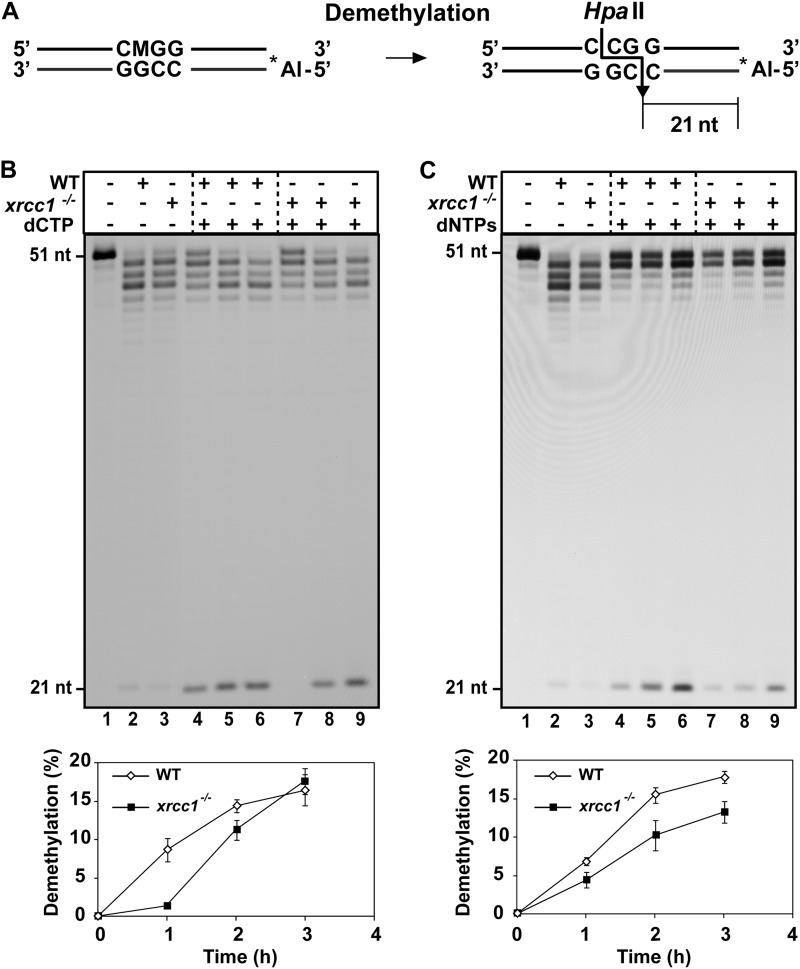
Cell extracts from xrcc1−/− mutants show a reduced capacity to complete DNA demethylation. A, schematic diagram of molecules used as DNA substrates in the demethylation assay. Double-stranded oligonucleotides contained a 5-meC (indicated as M) at an HpaII site on the upper strand. The Alexa Fluor-labeled 5′-end is denoted by an asterisk. The size of the 5′-end-labeled fragment generated after HpaII digestion of the fully demethylated products is indicated. B, purified ROS1 (70 nm) was incubated at 30 °C for 8 h with a double-stranded oligonucleotide substrate (20 nm). Reaction products were purified and incubated with 35 μg of cell-free extract from WT and xrcc1−/− mutant plants at 30 °C in a reaction mixture that contained either dCTP (B) or all four dNTPs (C). Reactions were stopped at different times (1, 2, and 3 h), and products were separated in a 12% denaturing polyacrylamide gel and detected by fluorescence scanning. The lower panels show the percentage of DNA demethylation in reactions with WT (white diamonds) or xrcc1−/− extracts (black squares). Values are mean ± S.E. from two independent experiments.